The Validation Control Accuracy test pertains to the Variants workflow, and statistically measures the agreement between the called variants in the control sample assembly (i.e., “validation control”) against a curated answer set for that reference material (i.e., “standard”). Example reference materials include those from HapMap/1000 Genomes Project participant NA12878 (Zook JM et al., 2014), available through the Genome in a Bottle Consortium and the National Institute of Standards and Technology (NIST).
In this workflow, SeqMan NGen is used to create a validation control assembly. For detailed information on creating the control assembly, see the SeqMan NGen help topic Create a SeqMan NGen Assembly for Validation Control Accuracy Testing. Once the validation control assembly is complete, it is imported into ArrayStar for analysis.
The method you follow for importing the assembly into ArrayStar depends on your operating system, whether or not you have moved the assembly, and whether the SeqMan NGen wizard is still open.
If SeqMan NGen and ArrayStar are on the same Windows machine and the SeqMan NGen “Project Report” screen is still open:

Option A (easiest):
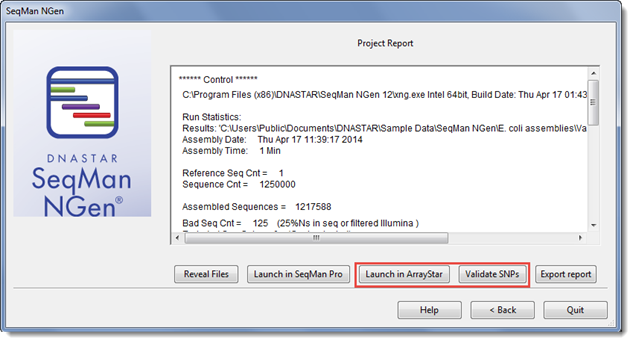
From the SeqMan NGen Project Report screen, click the Validate SNPs button. ArrayStar will launch the validation control sample. The associated VCF file will be loaded automatically and the validation statistics run.
Option B:
1) From within the SeqMan NGen Project Report screen, click the Launch in ArrayStar button. ArrayStar will launch with the project open. Check the Experiment List to make sure the VCF file associated with the control is also loaded.
2) To run the validation test, choose Statistics > Validation Control Accuracy. Specify the VCF as the Standard and choose which experiment to use as the Validation Control.
Note: Experiments settings have no effect on Validation Control Accuracy calculation, so nothing needs to be selected on the right side of the Statistics Parameters dialog.
3) Press Calculate to begin the Validation Control Accuracy calculation. Depending on the size of the project, a progress bar may be displayed.
In any other situation:
1) Launch ArrayStar manually (i.e., do not press the Launch in ArrayStar button from within SeqMan NGen).
2) Choose File > Import Experiments > Variants.
3) In the Add Experiments to Import screen, click Add SeqMan NGen .assembly folder and navigate to the Validation Control assembly. Note that the standard (VCF file) is loaded automatically when you load the control assembly.
4) Continue through the setup screens, leaving everything at the default values.
5) Once the setup wizard closes, choose Statistics > Validation Control Accuracy. In the Statistics Parameters dialog, specify the VCF file as the Standard and the .assembly experiment as the Validation Control.
Note: Experiments settings have no effect on Validation Control Accuracy calculation, so nothing needs to be selected on the right side of the Statistics Parameters dialog.
6) Press Calculate to begin the Validation Control Accuracy calculation. Depending on the size of the project, a progress bar may be displayed.

For information about the contents of the statistical report, see The Validation Control Accuracy Report.