Button Name
Icon
Description
Zoom In

Zooms in on the view horizontally to enlarge the Strategy View by 50% increments.
Zoom Out

Zooms out on the view horizontally to reduce the Strategy View by 50% increments.
Show Sequence Names

Displays the names of each constituent sequence in the Strategy View.
Hide Sequence Names

Hides the sequence names and compresses the arrows vertically, displaying more of the constituent sequences.
Paired End Data Display

Initially appears as a button with six colored arrows on it; it allows you to specify which reads should be displayed. When you click on it, a submenu of buttons opens. You may choose among the following options:

Shows all reads in the contig or contig scaffold.

Shows only paired reads.

Shows only reads that do not have a paired partner in that contig or contig scaffold.

The dark and light blue arrows icon shows paired reads located in different contigs whose assembly locations are consistent (dark blue) or could be made consistent (pale blue) with the Pair Specifier parameters

The red/orange arrows icon shows paired reads whose assembly locations are inconsistent with Pair Specifier parameters

Launches a dialog where you may customize which types of paired end data to display.
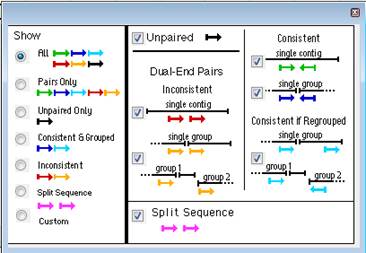
Using the radio buttons on the left, select any of the displays described above, or choose Custom. If you choose Custom, you may select items to be displayed using the checkboxes in the center and right-hand columns of the dialog.
Stranded coverage / Pair consistency tool


When available, this button toggles between displaying stranded coverage or pair consistency. This button is only available for certain RNA-Seq assemblies originating from SeqMan NGen.
Navigate tool

Click the compass rose icon to open a search dialog. Click the buttons to find the next or previous single-sequence Cov(erage) area, Feat(ure) or SNP. Clicking Next (or Prev) Cov causes the cursor to stop at both the beginnings and ends of single-sequence coverage areas.