Note: A contig that is part of a scaffold will have the scaffold number appended to the contig name:
The Alignment View displays the consensus sequence and the alignment of all sequences making up a contig at the nucleotide level. The consensus sequence for the selected contig is displayed at the top of the window, immediately beneath the scale. Constituent sequences are displayed in the bottom section. Translations and trace data can also be displayed in the Alignment View. The capitalization and color of residues convey additional information.
To access the Alignment View for a contig, double-click on the contig name in the Project Summary window, or select the contig name(s) and then select Contig > Alignment View. When you choose a contig scaffold or multiple contigs, each contig appears in its own Alignment View window.
Note: A contig that is part of a scaffold will have the scaffold number appended to the contig name:

•The standard Alignment View is used for projects without multi-sample data.
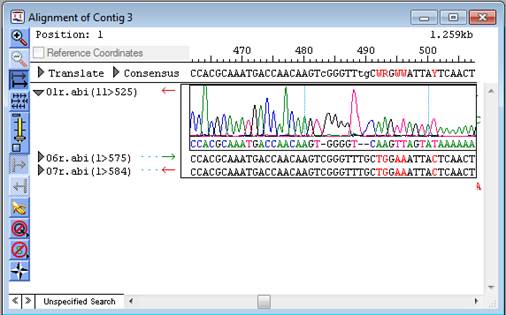
In addition to viewing the sequences, their corresponding traces or flowgrams (if available), the consensus sequence, and corresponding translations, the standard Alignment View allows you to edit the consensus and constituent sequences.
Note: If you are viewing a SeqMan NGen assembly in which DNA sequencing reads were assembled against an RNA reference sequence, the reference row in the Alignment View will contain U’s where the read rows have T’s. Though not actually mismatches, the T’s will appear in red, which indicates a “mismatch” in other circumstances. By contrast, the T/U equivalency is recognized in the Variant Table, and T’s there are not colored blue (which would indicate a variant).
•The enhanced Alignment View is used for projects with multi-sample data. See Multi-Sample Data in the Alignment View to learn about additional options available only in that version of the view.
Both versions of the Alignment View provide right-click command options, which are available by right-clicking on a sequence, rather than the name of the sequence.
|
Task |
Instructions |
Enabled for BAM? |
|
To select an entire sequence |
Click on the sequence, then right-click and choose Select All. |
Yes |
|
To cut, copy or clear the highlighted portion of the sequence |
Select some or all of the sequence, then right-click and choose Cut, Copy or Clear. Cut and Copy place the highlighted sequence on the clipboard, whereas Clear removes it without placing a copy the clipboard. |
Copy only |
|
To copy the name of the sequence |
Click on the sequence, then right-click and choose Copy Seq Name. |
Yes |
|
To paste the contents of the clipboard |
Click at the desired insertion point, then right-click and choose Paste. |
No |
|
To undo the last step |
Right-click and choose Undo to revert back one step. This command can be used to undo procedures such as Cut, Clear, Paste or Trim Ends. |
No |
|
To view trace or flowgram data in a separate window |
Click on a sequence, then right-click and choose Show Original Trace/Flowgram Data. If you click on the consensus sequence before using this command, SeqMan Pro will open separate windows for each sequence that is part of the consensus at the point of selection. |
No |
|
To display/hide all trace data chromatograms |
Right-click on any sequence that has associated trace data and choose Show All Traces or Hide All Traces. |
No |
|
To view sequence or contig information |
Click on a sequence, then right-click and choose Show Seq Info. If you click on the consensus sequence before using this command, SeqMan Pro will open separate windows for each sequence that is part of the consensus at the point of selection. To see contig information, click on the consensus, then right-click and choose Show Contig Info. |
Show Contig Info only |
|
To export and open the consensus in another Lasergene application |
Click on the consensus, then right-click and choose Send Consensus To. Choose from the list of available applications. The other Lasergene application will launch with the exported consensus already open. |
Yes |
|
To adjust endpoints for multiple sequences |
To trim from the 5’ end of a contig – Click before the first consensus base and drag the mouse to the right to select the ends of sequences you want to trim, then right-click and select Trim Ends. To trim from the 3’ end of a contig – Click after the last consensus base and drag the mouse to the left to select the ends of sequences you want to trim, then right-click and select Trim Ends. |
No |