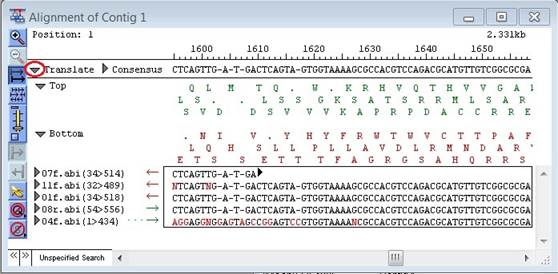
To view amino acid sequences for both strands of the consensus, click the triangle to the left of the word Translate. The amino acid sequences for the top and bottom strands will appear beneath the consensus sequence.

•Forward and reverse translations may be closed individually by clicking on the arrows to their left, labeled Top and Bottom.
•Gaps in the consensus sequence appear as spaces in the translation, while termination sites are shown as dots.
•Amino acids are displayed, by default, using 1-letter codes. To use 3-letter codes, un-check the box next to Use single letter protein codes found in the Editing & Color parameters.
•Translation is done using the current Genetic Code settings.
Note: An initial translation for a larger contig may take more time to compute than for subsequent viewings.