- If you have not yet downloaded and extracted the tutorial data, click here to download it. Then decompress (unzip) the file archive using the method of your choice.
- If a MegAlign Pro project is already open, choose File > New Alignment Project > Blank Document. Otherwise, launch MegAlign Pro.
- Choose File > Add Sequences End-to-end, or click on the Add sequences end-to-end in project tool (
).
- Click the Add button, then navigate to and open the tutorial data folder Rp1 exon groups. The nucleotide files in this folder represent the RP1 exons for several mammal species. Each exon consists of 3-4 segments.
- If needed, click either once or twice on the file name header to ensure the sequences are in ascending alphabetical order, as in the image below.
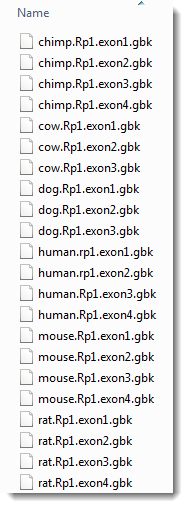
- Use Shift+click to select only the four "chimp" segments, and then click Open. In the Add Sequences End-to-end dialog, the files will appear in ascending alphabetical order.
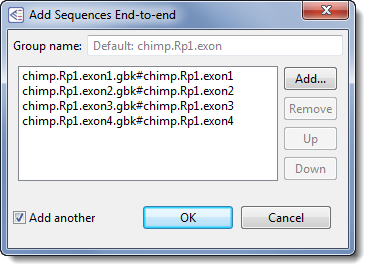
- In the Group name text box, replace the default name with Chimp, and then click OK.
- Follow steps 3-5 four more times to create separate groups for the Cow, Dog, Human, and Mouse sequences. In each case, replace the generic Group name with the informal species name.
- For Group 6, add the rat sequences and replace the default name with Rat, but do not yet click OK. For demonstration purposes, you will deliberately be ordering these sequences incorrectly.
- Select the "exon1" sequence by clicking on it once, and then click three times on the Down button until the sequence appears at the bottom of the list. Similarly, select the "exon4" sequence, the click twice on the Up button until the sequence appears at the top of the list.
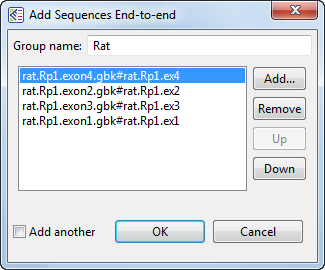
- Remove the checkmark next to Add another, and then click OK.
Looking at the Overview, observe that each species is represented by multiple gray blocks of different lengths. Blue dividers show where one segment ends and another begins.
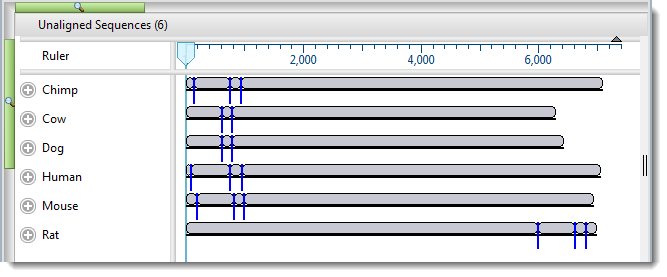
- To make the blocks appear smaller or larger, use the horizontal and/or vertical zoom sliders ().
- Select Align > Align Using MUSCLE to align the sequences using the MUSCLE algorithm. The alignment will likely take several minutes to complete.
- While the alignment is being performed, try using the View > Console command. This opens a log showing the progress of the alignment. Before proceeding, close the Console by clicking the ‘x’ in its tab.
- Use File > Save to save the project under the name RP1 project.msa.
Proceed to Part B: Replace a sequence in the Overview and view the phylogenetic tree.
Need more help with this?
Contact DNASTAR



 ).
).