In this part of the tutorial, you will explore how to view and change parameters prior to performing a multiple sequence alignment.
- Choose Align > Align Using Mauve and align the sequences using the default parameters. Because this is a genomic alignment, the alignment will take longer than those done in the first two tutorials (approximately 5-10 minutes).
After genomic sequence alignment with Mauve, MegAlign Pro displays genomes in the Overview as Locally Co-linear Blocks, usually abbreviated to "blocks" or "LCBs." Blocks represent multiple-aligned sections of at least two of the current set of genomic sequences. Discontinuity in numbering is displayed as a break between blocks. Connecting lines show the correspondence between blocks. Blocks hanging below the black line, signify that the hanging blocks are on the opposite strand compared to the other blocks.
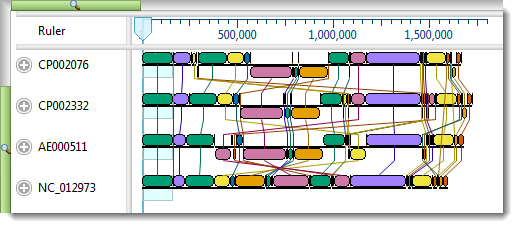
The default settings used in creating this alignment are suitable for most projects, including this one. However, you will make a change to Mauve’s Seed weight parameter in order to see how this affects the alignment results.
In Mauve, Seed weight is the minimum weight of the seed pattern used during the first pass of a multiple alignment. When aligning divergent genomes, lower seed weights may provide better sensitivity. However, if the value is set too low, the result will be reduced sensitivity. In the following steps of this tutorial, you will lower the Seed weight slightly, from the 15 (the default) to 13.
- Choose Align > Align with Options. This dialog allows you to change methods and/or parameters. Note that Mauve is already selected in the menu at the top of the dialog, since it is the last method used.
- By default, the Default seed weight box is checked, meaning that the optimal seed weight (number of matches required per k-tuple) is calculated automatically. Uncheck the box and either use the slider to change the Seed weight to 13, or type 13 in the Seed weight text box.

- Choose Align to perform the Mauve alignment again using the selected parameters.
Proceed to Part C: View a genomic alignment in the Overview.
Need more help with this?
Contact DNASTAR


