- If you have not yet downloaded and extracted the tutorial data, click here to download it. Then decompress (unzip) the file archive using the method of your choice.
- If a MegAlign Pro project is already open, choose File > New Alignment Project. Otherwise, launch MegAlign Pro.
- Outside MegAlign Pro, use the Windows Explorer or Macintosh Finder to locate and open the tutorial data folder H. pylori genomes. Arrange and size the file explorer and the MegAlign Pro window so that you can see both simultaneously.
- In the file explorer, use Shift+click to select the four files.
- Use the mouse to drag & drop the selected files onto any "white space" in the Overview or Sequences view.
The sequences appear in the Overview as solid gray blocks of unequal length, indicating that the sequences have not yet been aligned.
- Use your mouse to drag and drop the sequence names (on the left side of the window) so they appear in the same order as in the image below. This will ensure that your block colors and positions match those referenced in later sections of this tutorial.
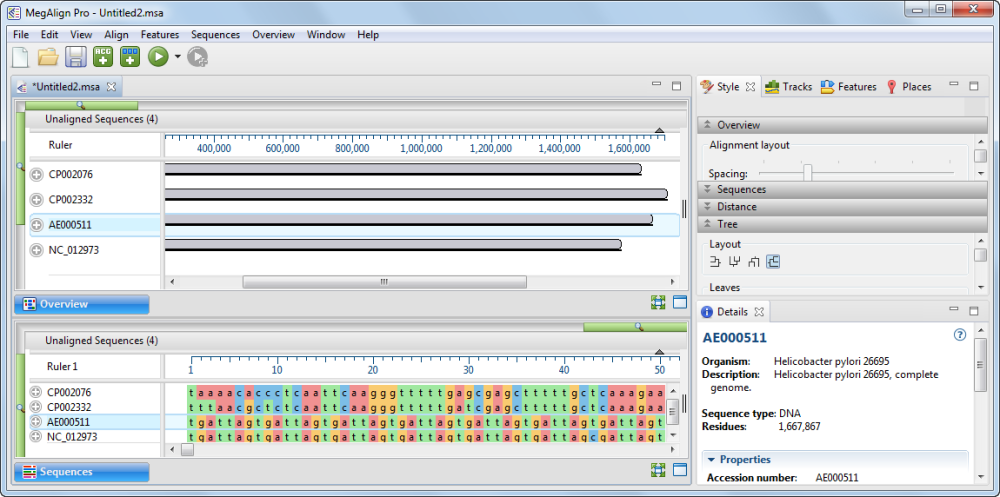
- Use File > Save to save the project under the name H. pylori project.msa.
Proceed to Part B: Perform a Mauve alignment using modified parameters.
Need more help with this?
Contact DNASTAR


