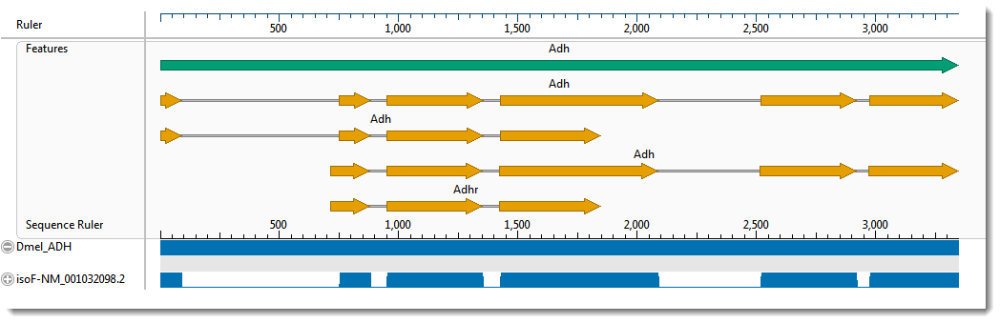
Since the Local pairwise alignment in Part B was not an improvement over the multiple alignments in Part A, you will now try a Global pairwise alignment. This type of alignment forces the ends of both sequences to be aligned.
- Click the gear icon on the Pairwise view toolbar and select Global: Needleman-Wunsch from the “Using” drop-down menu. Then press Align.
- After aligning, the display options that you set above remain in effect, but the sequences’ detail tracks will collapse. Expand the DmeI_ADH track again by clicking on its plus sign.
- To see the “big picture,” zoom all the way out using the green “zoom slider” above the view. The pairwise alignment now appears similar to the Overview. You can see that the global alignment using isoF matches Dmel_ADH’s annotations. (Click on the image to expand it.)
MegAlign Pro allows you to open additional pairwise views so that you can compare alignments with different settings, or even different pairs of sequences. You can use this to compare any of the other three mRNA sequences to Dmel_ADH, while leaving the previous alignment untouched.
- Clone the current Pairwise view using the ”Clone this view” tool situated at the far right side of the toolbar. The two views will start out the same, but they are completely independent.
- Select isoE-NM001032099.2 as the query sequence, keeping the reference sequence set to Dmel_ADH. The new Global pairwise alignment now gives the expected results.
This is the end of the tutorial.
Need more help with this?
Contact DNASTAR