You can view a portion of a sequence as folded RNA. GeneQuest uses the Vienna RNA folding procedure, taken from Zuker’s optimal RNA folding method, to fold the sense strand of selected DNA regions as RNA.
Use the Range selector to select a portion of sequence. The selection must be 1500 bases or less in length. Then choose Analysis > Fold As RNA. The folding procedure may take up to several minutes.
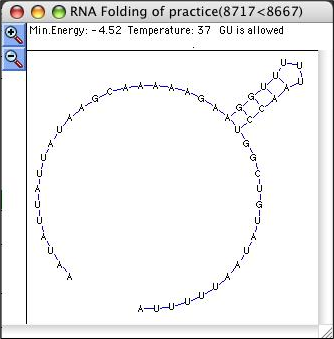
The folding model is an interactive two-dimensional plot. At the top of the RNA folding window, GeneQuest lists the minimum energy (in Kcal/mol) of the structure, the temperature used in folding and the status of G-U pairing. Use Zoom In (

Editing RNA folding parameters:
Parameters can be changed either before or after folding RNA. To set parameters before folding the selection, hold down the Alt key while selecting Analysis > Fold As RNA. To alter the parameters after viewing the folded RNA, select Options > Folding Parameters. The command is hidden unless the RNA folding window is the active window. In both cases, the following dialog opens.
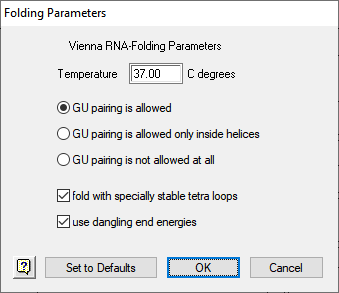
- Enter the Temperature that you want to be used in the folding algorithm. The default is 37°C. Altering temperature will modify the nearest neighbor free energy values (Frier et al., 1986) used to calculate the minimal free energy of the folded structure.
- The next three buttons determine the state of guanine to uracil base pairing. You can allow G-U pairing everywhere, allow it only inside helical structures, or forbid it entirely. Allowing G-U everywhere will increase the complexity of secondary structures and forbidding it will decrease complexity.
- Checking either of the two checkboxes, fold with specially stable tetra loops or use dangling end energies, will cause GeneQuest to use alternate nearest neighbor free energy values while within these respective structures.
Need more help with this?
Contact DNASTAR


