The Secondary Structure – Garnier-Robson track , from Garnier et al. (1978), Garnier and Robson (1990), and Garnier, Gibrat and Robson (1996), predicts protein structure from the amino acid sequence. The method examines the propensity of a given residue to exist in a certain structure. It is a statistical approach which uses examination of known crystallographic structures to define regions as α-helical (H), ß-pleated sheet (E - extended chains), ß-turns (T) and coil ( C ).
To apply this track to the sequence:
In the Tracks panel, expand Secondary Structure and check the box next to Garnier-Robson. The track will now be visible in the Analysis view
To edit track options:
Select the track in the Tracks panel. Open the Track Options section, which appears as follows:
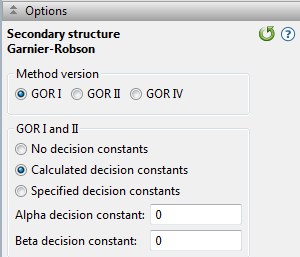
Specify which version of the GOR method you wish to use: GOR I, GOR II or GOR IV. GOR II uses the same algorithm as GOR I, but is based on updated propensity tables. GOR IV does not predict turns, but only predicts the probability of areas being helices, sheets or coils. GOR IV is the most modern secondary structure prediction method in Protean 3D.
If you choose GOR I or GOR II, you may select from among three options related to the decision constant:
- No Decision Constants - if you do not want Protean 3D to make assumptions about the global α-helix and ß-sheet content.
- Calculated Decision Constants - to use computer-derived constants from global α-helix and ß-sheet probabilities. Constants are based on three protein classes: proteins with less than 20%, 20-50%, and greater than 50% α-helix or ß-sheet.
- Specified Decision Constants - if you have prior knowledge of circular dichroism data. If you select this button, you need to input values for Alpha Decision Constant and Beta Decision Constant.
Click 
Need more help with this?
Contact DNASTAR


