The Transmembrane – TopPred track utilizes hydropathy analysis to propose probable transmembrane regions in a protein sequence, using the methods of von Heijne, 1992, and Claros and von Heijne, 1994. Possible membrane-protein topologies are considered and ranked according to the “positive-inside rule,” an observation that the cytoplasmic parts of membrane proteins more frequently contain positively charged residues compared to non-cytoplasmic parts. Additional considerations are made for eukaryote proteins, including the net charge difference of residues flanking the N-terminal transmembrane segment.
To apply this track to the sequence:
In the Tracks panel, expand Transmembrane and check the box next to TopPred. The track will now be visible in the Analysis view
To edit track options:
Select the track in the Tracks panel. Open the Track Options section, which appears as follows:
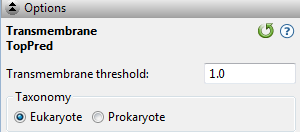
- Transmembrane threshold – Regions with a maximum hydrophobicity score equal to or greater than this threshold will be reported as transmembrane regions, as long as they are not adjacent to, nor overlap with, other transmembrane regions. A setting of 0.6 will cause Protean 3D to report all putative regions, while a threshold of 1.0 will cause the application to be more selective and only report regions with a higher probability of being transmembrane regions. The default is 1.0.
- Taxonomy – Select the taxonomy of the organism from which the sequence is derived. The selection influences Protean 3D’s prediction of whether a region is cytoplasmic or extracellular.
Click 
Need more help with this?
Contact DNASTAR


