The Annotation Results window displays putative matches between SeqBuilder Pro’s plasmid Feature Library and one or more query sequences, usually vector sequences from a cloning project. If the query sequence lacks some/all of the located features from the plasmid Feature Library, or contains incomplete or obsolete versions of certain features, this window allows you to copy features from the database to the query sequence. To open the window, select one or more sequences from the Project window and then choose Features > Annotate Sequence.
This topic describes the layout of the window. To learn how to perform a search, evaluate putative feature matches and add them to the query sequence, refer to Perform the auto-annotation procedure.

Components of the Annotation Results window are described below from top to bottom. However, note that the selection made in the lowermost pane determines the contents of all other panes.
After you have examined putative feature matches in the table, use the Accept Checked tool (

The header area shows how many matching features were found, and how many were unique.

The Matched Feature pane near the top of the window shows the full width of the selected feature in the style specified in the Feature Library Manager. If no style has been specified, it is instead shown in the default style.

The Match Quality pane displays an alignment histogram that uses the query vector’s coordinate system to show where a plasmid Feature Library feature matches the query sequence. The top bar represents the feature from the plasmid database, while the bottom bar shows the location of that match on the query. Gaps appear as empty space, mismatches are displayed in yellow, and matches appear in green.

The Matched Feature/Context Features pane uses the query vector’s coordinate system to show features that overlap or are near to the feature selected in the table. Gray vertical lines at the endpoints of the feature can be used to find features with identical or near-identical endpoints. Imperfect matches can still represent duplicates. For corroborating evidence, look for significant overlap in features, as well as text field matches.

The Matched feature and Putative feature panes just above the “annotation match table” lets you compare text information from the feature in the query sequence (on the right) and the putative plasmid Feature Library match (on the left). If the query sequence does not already contain the putative feature (i.e., the row in the table below is checked by default), the right pane will say: “No putative duplicate found.”

The Annotation match table in the lower half of the window consists of a list of annotations that match the query sequence at the specified thresholds for match and coverage percentage. To sort entries alphanumerically based on a particular column, click on the corresponding column header.
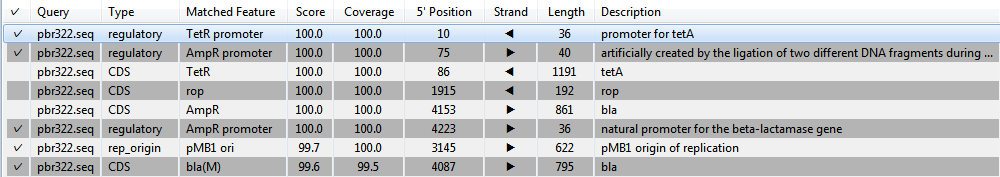
The table columns are described below:
| Column name | Description |
|---|---|
 |
A check mark at the left of a row signifies a unique annotation: it exists in the plasmid Feature Library, but not in the query sequence. If multiple versions of an annotation match the query, only the best match is checked, by default. An unchecked row signifies either a non-unique annotation or a duplicate annotation where one of the other duplicates was a better match. The leftmost table cell for each row is used to add or remove these check marks manually. When you click on the cell, a menu opens with the following choices:
|
| Query | The name of the query sequence that the plasmid Feature Library match pertains to. |
| Type | Feature type (e.g., CDS, regulatory, rep_origin, etc.) |
| Matched Feature | Name of the feature for which a match was found. |
| Score | Likelihood that the plasmid database feature is a match for the query sequence, based on calculations made within SeqMan NGen. |
| Coverage | (Length of the plasmid database feature / length of the putative match in the query sequence) x 100% |
| 5’ Position | 5’ Position of the plasmid database feature on the query sequence’s coordinate system. |
| Strand | Contains an arrow showing the strand direction, left (  ) or right ( ) or right (  ). ). |
| Length | Length of the plasmid database feature. |
| Description | Description of the plasmid database feature. |
Need more help with this?
Contact DNASTAR


