To join two or more sequences end-to-end:
- Choose Tools > Concatenate. The Project window opens with the Project and Options tabs active.
- In the Project tab place the sequences you wish to join into the Sequences folder. To add sequences to a folder, select the folder, then right-click it and choose Import. Select the desired sequence files, and press Open. Use Shift+click or Ctrl/Cmd+click to select two or more sequences.
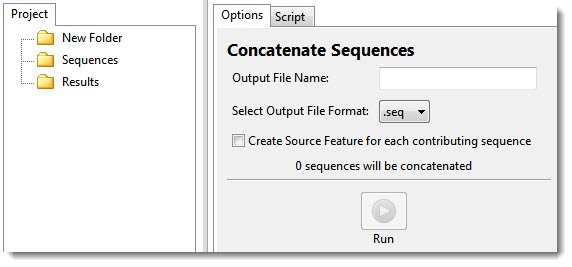
In the Options tab on the right, the number of sequences selected on the left appears in the message: “‘n’ sequences will be concatenated.’
- In the Options tab, type a name (and extension, if desired) in the Output File Name text box.
- Select an output file format from the drop-down menu. Choices include .gbk (preserves annotations) or .fas (no annotations).
- If desired, check the Create Source Feature for each contributing sequence box. Checking this box creates a “source” feature for each sequence that contributes to the concatenated sequence. This can assist you in keeping track of where each sequence originated.
- Press the Run button.
The new sequence appears in the Project tab’s Results folder under the name specified. No file extension is appended to the name unless one was specified in the Output File Name box.
- (optional) If you wish to see the script that was used to run this process, press the Script tab.
- (optional) If you want to see the history of the run, including the location of the output file(s), press the History tab.
Example:
Input sequence 1: GGAGTCTCCC
Input sequence 2: ATTGATTACA
Output sequence: GGAGTCTCCCATTGATTACA
Need more help with this?
Contact DNASTAR


