To create a primer pair for a region of interest on the sequence to use in TA cloning:
- If you have not yet download and extracted the tutorial data, click here to download it. Then decompress (unzip) the file archive using the method of your choice.
- Use File > Open to open the demo file Tn5w.sbd. The file opens in the Linear view.
- Click on the neomycin/kanamycin resistance feature labeled aminoglycoside-3’-O-phosphotransferase to select that range of sequence.
- Select Priming > Create Primer Pairs. The Create Primer Pairs dialog appears.
- Click the triangles next to Conditions and Primer Characteristics to expand those sections.
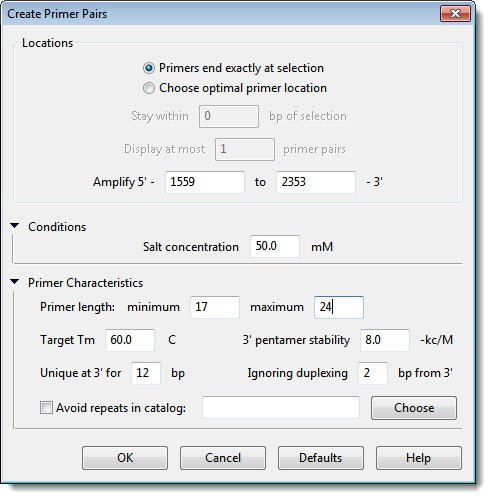
In most cases, you will not need to change the settings in this dialog. For the purposes of this tutorial, however, you are going to deliberately make a change such that the default Primer length values will not yield optimal results.
- Adjust the Target Tm to 55.0 C. Leave the rest of the values as they are, and then click OK.
SeqBuilder Pro will choose the best primer pair with the characteristics specified that lies within the selected region. The upper view of the SeqBuilder document is now the Primer Design view, focused on the forward strand of the primer. The lower view is the Primers view.
- If necessary, drag the dividing bar between the Primer Design and Primers views so that you can see all three sections of the Primer Design View: Residue, Mispriming and Alternate Pairs.

In the Primer Design view (upper pane), notice that, in the Mispriming section, the Most stable dimer and Most stable pair dimer are both labeled “BAD!” This indicates that the final pentamer value of the dimer exceeds the 3’ pentamer stability threshold defined in the Primer Characteristics section of the Create Primer Pairs dialog. In general, you should avoid using primers with dimers or hairpins labeled “BAD!” unless you have experimental evidence that they will function as desired, or unless you have no other option.
- In the Primers view (lower pane) the single primer pair located by your search is displayed. Click the plus sign next to Set 1 to expand the primer pair and view each individual primer within the pair. Notice that the melting temperature for the Forward primer is lower than the Target Tm of 55°C.

- In the Primer Design view (upper pane), select the codon change mode by clicking on the Introduce Codon Change/Mutation button (
) near the top left of the view. Notice that the cursor changes to an arrow, and that each codon is highlighted as you hover over the primer sequence.
- Introduce a codon change by clicking on the TTG triplet in the primer, located at 1577-1579 of your sequence. From the list of codons given, select CTA.

In the sequence area, the two changes to the primer sequence, and the corresponding changes to the translation, are shown in magenta in lower case.

(In the Primers view, these edits appear in lower case, but without any color change).
In the mispriming section, note the good news that the dimers/hairpins have either disappeared or are no longer marked “BAD!.” The bad news is that the header shows the Tm has now dropped further from the target temperature of 55.0°C, moving from 50.8°C to 44.1°C.
- Click the Selection mode tool (
) to exit from the current “codon change” mode.
- Referring to both the Tm in the header, and the Mispriming section at the bottom of the view, drag the triangle on the 3’ (right) end of the primer sequence to the right. Increase the length of the currently 22 bp primer, stopping when these two conditions are met: 1) the Tm is close to 55.0°C, 2) Mispriming contains no items labeled “BAD!.” This occurs when the primer is 31 bp in length and the Tm is 55.3°C.
- In the Primers view, name the modified pair by clicking on Pair 1 in the Name column and typing 55 degree pair.
- Save the project with any name and in any desired location using File > Save As.
The saved project is used in the tutorial Try it! – Use a PCR insert in TA cloning.
Need more help with this?
Contact DNASTAR



 ) near the top left of the view. Notice that the cursor changes to an arrow, and that each codon is highlighted as you hover over the primer sequence.
) near the top left of the view. Notice that the cursor changes to an arrow, and that each codon is highlighted as you hover over the primer sequence. ) to exit from the current “codon change” mode.
) to exit from the current “codon change” mode.