In Part A of this tutorial, you set up and ran a templated RNA-Seq assembly using SeqMan NGen. In this part of the tutorial, you will analyze the assembly results using ArrayStar’s advanced filtering functionality to create a set of genes that relate to the flagella structure.
- Choose Filter > Filter All to open the advanced Filter dialog.
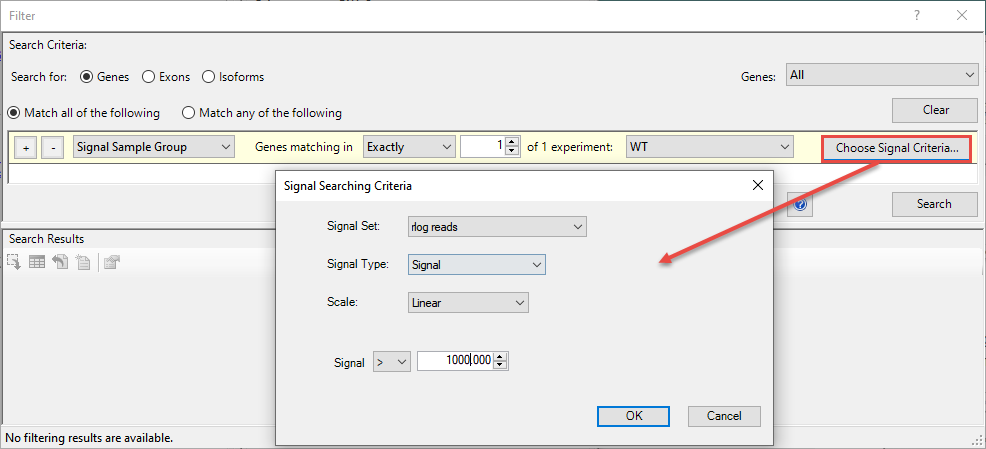
- In the yellow row, select options to match the image below. Note that you must press the Choose Signal Criteria button to open the pop-up dialog shown in the image. In the pop-up, change the Scale to Linear and the Signal to > 1000 (note greater-than sign). Press OK to close the pop-up.
- Click the plus sign (+) to the left of the yellow row to add a second row. In the new row, set up options to match the image below. In the Choose Signal Criteria pop-up, change the Scale to Linear and the Signal to < 100. Press OK to close the pop-up.
Press Search. This two-filter combination results in approximately 25-35 genes.
- Press the Select and show results in Gene table tool.

Observe that most of the genes that met the filter thresholds are fli and flg genes. These genes are involved in producing flagella in E. coli and are known to be regulated by the transcription factor encoded by the flhC and flhD genes. The che, tap and tar genes are involved in chemotaxis and are also known to be regulated by FlhCD.
- Use the Quick filter tool in the Gene Table header to Show all genes.
- Sort by the adjusted p-values from smallest to largest by clicking either of the p-value column headers.

Note that there are approximately 40-60 genes where the adjusted p-value is zero for both flhC and flhD. These include the genes found by filtering (still highlighted in blue), as well as additional flagella and chemotaxis related genes.

This marks the end of this tutorial.
Need more help with this?
Contact DNASTAR