SeqNinja allows you to substitute the values of settings into single- and double-quoted strings. You can substitute the value of any setting, including built-in settings, custom settings, and command-line arguments. When you run the script, SeqNinja will replace the name of the setting with its value. For example, a string (e.g., a file path) or a numeric setting (e.g., an integer) will be used in place of the setting name. This functionality allows the same script to be reused with different data and allows the script to be moved along with its data.
Substitution involves several steps, each of which is discussed below.
Step 1 – Enabling substitution:
Before substitution is used in a script, you must first enable “substitution” in that script. In the Visual view, this is easily done by adding the setting called Enable Substitution of Setting Values anywhere in the script above the substitution.

To enable substitution, the text box on the right must contain true, as shown in the image above. If it instead says false, overwrite the text with true.
Step 2 – Defining a setting:
Settings can be defined…
- in the script.
- as an argument typed into the Run with Options dialog.
- implicitly (see the “Tip,” below, for an example).
To define a setting in the script:
In an open script, click on the Settings tool User Setting Prefix. The following step is added to the script:

By default, the drop-down menu on the left is set to user. To differentiate this custom setting from other custom user: settings, you may type additional text to the right of the colon, if desired. For example, user:tags, user:fasta, user:genbank.
In the text box on the right, enter the information you want to substitute for the setting when the script is run (e.g., a file path, integer, etc.). Substitution can occur in numeric expressions or in strings. Strings are sequences of characters (e.g., a file path) between pairs of single or double quotes.
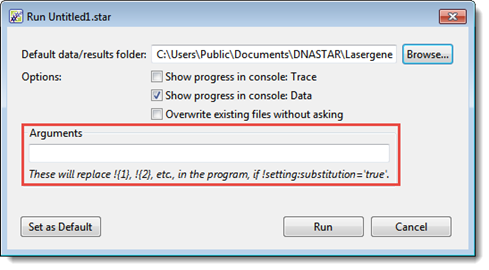
To define a setting as an argument in the Run with Options dialog:
SeqNinja lets you define a custom setting as a script argument in the Run with Options dialog.
- After setting up the script as desired, choose Run > Run with Options.
- Type one or more arguments into the Arguments box. Multiple arguments should be separated by a space. Note that an argument can only define settings whose name is a positive integer.
Example: You set up a script that deletes the first ‘n’ bases of a sequence. The script uses !${1} to refer to the sequence file, and !${2} to specify the number of bases (‘n’). In this example, you could use the argumentsmyfile.fastafor !{1} and42for !{2}. Therefore, you would enter the textmyfile.fasta 42into the Arguments box.
3) Reference the setting in the script using !{1}, !{2}, and so forth. If you are creating a script in the Visual view, there is no specific step in which the setting is referenced. Simply type the substitution text into any appropriate text box.
Step 3 – Specifying substitution:
Curly braces are used to substitute the value of a setting in a string or numeric expression.
Examples:
Examples related to defining custom settings are shown in the table below.
When you create a script in the Visual view by adding “steps,” a corresponding piece of scripting language is added to the script . This underlying script can be seen by switching to the Text view, where it can also be edited. You can then review the steps in the Visual view, if desired, before running the script.
For instance, if you added the script in the second row (“Sequence literals”) in the Text view and then looked at it in the Visual view, it would appear as:

| Context | Example | Comment | |
|---|---|---|---|
| Paths | "out.fap" = "!{program:dir}/adh4_mouse.pro" |
||
| Sequence literals | "out.fasta" = "!{1}" + "in.fasta" |
In this example, the user passes a sequence of residues as the first command-line argument. For literals defined in the script, it’s easier just to use a sequence variable: $INSERTION = "NNNNNNNN" "out.fasta" = $INSERTION + "in.fasta" |
|
| Patterns | "out.fasta" = collect('!{script:dir}/genomes/Vibrio*.gbk') |
||
| Sequence index | "out.fasta" = "454.fasta"#"${1}" |
||
| Settings | !mydir="C:\Users\norville" !mydata="!{mydir}/data" !myresults="!{mydir}/results" |
||
| Integers | out.fasta = in.gb ( %!{1} > %!{2} ) |
This allows using integers passed on the command line in numeric expressions. For values defined in the script, it’s easier just to use variable integers: %A = 42 %B = 111 out.fasta = in.gb( %A > %B ) |
|
| Single-quoted strings | print( 'File: !{1}' ) |
Need more help with this?
Contact DNASTAR