In the SNP Table, some SNP Annotation columns feature a color display.
The Amino Acid Change, Called Seq, Classification, Genotype and Type columns:
By default, the following color scheme is used in all of these columns:

For example, red text denotes a non-synonymous change, while white text (i.e., no text visible in that cell) indicates a lack of coverage at that position.
If you would like to change the color scheme used in these columns, for either display or print purposes, follow the procedure described in Edit Color Settings.
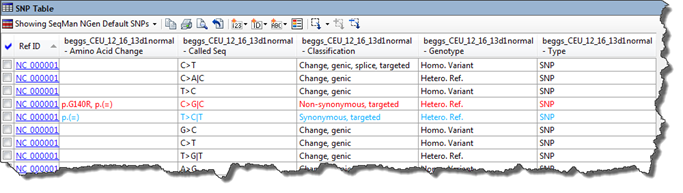
The Called Seq column:
In the Called Seq column, gray is used to denote a sample containing a SNP at the same position where other samples contain the reference base.
Example: In SeqMan NGen, use exome data from a mother, a father and their son to create three separate assemblies. The workflow used for this specifies that variants should be calculated.
In ArrayStar, create a Variants project and loaded the three assemblies. Open the SNP Table and add a Called Seq column for each family member. To find a family member that contains a SNP at the same position where other family members have the reference base, look for text in gray.
