There are several ways to import annotations during project setup.
If you are following the Variants workflow, variant annotations from a VCF file can be imported using the Add File button in the Set Up Preprocessing screen. In addition, VCF files with sample information can be loaded into a project as a separate experiment; for files with multiple samples, each sample will be loaded separately. ArrayStar determines whether the VCF file has sample information by looking for the “FORMAT” column in the header line according to the VCF specification. Columns to the right of FORMAT should contain sample information. If you have a validation control and wish to run accuracy statistics on that experiment, the VCF file for that experiment must contain sample information. If that VCF was specified as part of the assembly, it is automatically loaded into the ArrayStar project.
If you are following the Microarray workflow, the Import Annotations step of the Project Setup Wizard allows you to import available annotations for your data. This step of the wizard varies depending on the type of data files that have been loaded. For details, see:
o Importing Annotations for Affymetrix Data
o Importing Annotations for NimbleGen Data
o Importing Annotations for All Other Microarray Types
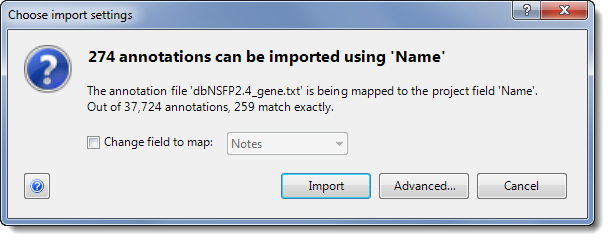
When importing annotations, ArrayStar only loads those which are associated with genes in the project. For information on the resulting dialog (“Choose import settings”), see File Import Mapping.

Note: If you forget to import annotations during this step of project setup, you can also import annotations after project setup is complete.