The Min/Max Cap transformation caps all of the values in your project at a defined minimum or maximum value. This transformation enables you to treat as equal all values which fall below or rise above a level where they may not be valid. For example, if all of the data that falls below a linear level of 100 is drowned out by noise on the array or in the scanning process, you may use a minimum cap to set all of these to 100 in order to ignore changes below this level.
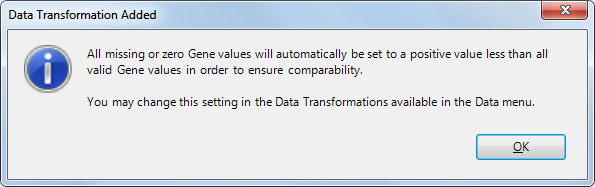
When a new project is created, ArrayStar automatically provides, by default, a minimum value for all genes that have zero or missing expression values. To do this, ArrayStar finds the lowest positive value in all of the experiments, then uses half of that value in lieu of any zero or missing values. This calculation does not alter any positive values. If this Min/Max Cap transformation were not performed, it would be possible to filter on Fold Change in the Advanced Filters dialog, but still miss any genes that were turned "off" in the experiment.
Notification of this automated data transformation appears during the Set Up Preprocessing step of new projects:

To apply the Min/Max Cap transformation to an existing ArrayStar project, open the Edit Data Transformations dialog by selecting Data > Edit Data Transformations from the main menu. You may also apply the Min/Max Cap transformation in the Set Up Preprocessing step of the Project Setup Wizard.
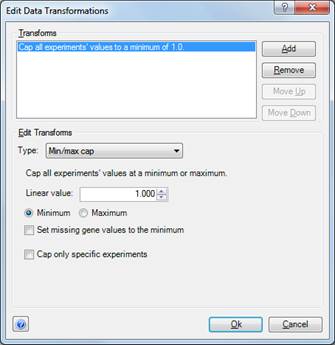
•Click the Add button to populate the dialog.
•From the Type drop-down menu, select Min/max cap.
•Next to Linear value, type in the desired number for the cap. Then select the Minimum or Maximum radio button to denote which of these your cap number represents.
•If Set missing gene values to the minimum is unchecked, only data points which actually have a valid value which is less than the Minimum are capped (set to the Minimum). If checked, all missing values are set to the Minimum as well. This may dramatically increase how many data points are displayed in some views with some data sets.
•If Cap only specific experiments is unchecked, the Min/max cap transformation is performed on all experiments in the project. If checked, you may specify which experiments' values are affected.
Default values vary depending upon when the min/max cap transformation is added:
|
When added |
Minimum/Maximum? |
Linear value |
Set missing gene values to the minimum |
Cap only specific experiments |
|
During data import |
Minimum |
Auto-Calc |
checked |
checked |
|
At any other time |
Minimum |
1.0 |
unchecked |
unchecked |