 next to the
Quick Search field
and selecting Advanced Filtering.
next to the
Quick Search field
and selecting Advanced Filtering. The Filtering dialog can be accessed through the Filter menu
(see ArrayStar Menu
Commands) or by clicking  next to the
Quick Search field
and selecting Advanced Filtering.
next to the
Quick Search field
and selecting Advanced Filtering.
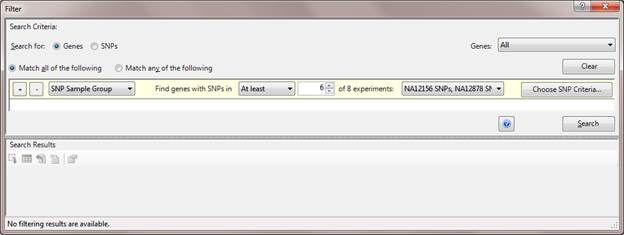
The Advanced Filtering dialog has been equipped with extensive options to allow flexibility in searching for variations of interest amongst the sample panel. The following are examples of filters that can be set up using the Advanced Filtering dialog:
•Search only for items (e.g., SNPs, genes, exons, isoforms) that are present in a particular experiment.
•Search for items that are present in ≥ X% of the samples.
•Search for items that are present in ≥ X% of Sample Set A, but not present in > Y% of Sample Set B.
•Search for items that meet specified statistical criteria.
Setting up a filter:
The Search for field defines what signal values will be filtered for the search. The subset of available options will depend on the type of project you are working with:
•Microarray projects - Genes
•CNV and RNA-Seq projects – Genes, Exons or Isoforms
•ChIP-Seq projects - Genes, IP Peaks, IP Fragments, Exons or Isoforms
•miRNA projects - Genes, IP Peaks, IP Fragments, Exons or Isoforms
•Variants projects – SNPs, or the Genes containing those SNPs
Select the radio button next to Match all of the following to indicate that all of the criteria you specify must be met. Select Match any of the following to indicate that only one of the criteria must be met for the item to be included in your search results.
The field chosen in the Search for area defines which items will be searched for your criteria. Select All to search all instances of that item or Selected to search only the currently selected items. Or choose any of your saved sets to search only the items in the specified set.
Click the plus sign to add another row of search criteria. Click the minus sign to remove a row from the list.
A wide variety of search criteria are available. The set of text boxes and drop-down menus on each row vary depending upon the experiment type and the selection made in the Search for field. Use the pull-down menus and numerical text boxes to enter your filtering criteria.
In the left-most drop-down menu, available options will include a subset of the following: Annotation Text, Expression Level/Signal, Sample Group, SNP Search, Fold Change, Gene Classification/Ontology, and Statistics.
Returning the filter to default values:
If desired, click the Clear button at any time to forget your changes and return all dialog settings to their default values.
Performing the search and working with search results:
After entering your criteria, click the Search button in the bottom right of the window. Your results will be displayed in the Search Results pane beneath the window. See Working with Advanced Filtering Search Results for more information.
Viewing “hidden” search results:
The columns displayed in your Search Results pane, shown in
the bottom half of the Advanced Filtering window, are controlled by your Gene
Information Settings. In order to view fold change, statistics, and signal
values for the genes found in your search results, click the  button from the toolbar, and then go
to Data > Show Gene Table. Once you are in the Gene Table, it may be
necessary to click the
button from the toolbar, and then go
to Data > Show Gene Table. Once you are in the Gene Table, it may be
necessary to click the  tool to display only your search
results.
tool to display only your search
results.