Menu
Command
Description
File
New Project
Use this option or press Ctrl + N to create a new, unnamed project. By default, an empty Experiment List will be opened.
Open Project
Use this option or press Ctrl + O to open an existing ArrayStar (.astar or .dmaproj) project file.
Close Project
Use this option or press Ctrl+Shift+W to close the current project. If the project has never been saved, or has been changed since the last save, you will be prompted to save it.
Save Project
Use this option to save the current project. If you are saving an existing project, ArrayStar writes over the previous version. If you want to preserve the earlier version, select File > Save Project As to save the project under a different name.
Save Project As
Use this option to store a copy of the current project under a new name. The original project will be closed under its original name as it appeared when last saved. You can also use this command to rename a document or to save a document to a different location.
Import Experiments
Choose a subcommand from this option to import data files to either a new or existing project through the Project Setup Wizard. For additional information regarding the file types that ArrayStar supports, please see the Supported File Types section.
|
Sub-command |
Workflow |
|
SNP |
Compare SNPs from genomes and exomes; access the Variant Annotation Database. |
|
CNV |
Compare genomic copy number variations (CNVs); genes derived from exons |
|
RNA-Seq |
Compare gene expression and ontology; genes derived from isoforms (RNA-Seq) |
|
ChIP-Seq |
Compare transcription factor binding sites (ChIP-Seq) |
|
miRNA |
Identify micro-RNAs |
|
Microarray |
Compare gene expression and ontology |
Import Annotations
Use this option to import an annotation file into your current project. This option is only enabled after you have imported data into your project. If you import a .txt file, the Data Import Wizard will be launched.
Import Set
This option is only available in the Set List view. Use this option to import a gene set (.txt file) into your project.
Import Special
When visible at all, this command is disabled. The command acts as a File Menu placeholder for functional commands accessible from within other views or tables: File > Import Experiments, File > Import Annotations, and File > Import Set.
Export Selected Experiment
The option is only available in the Experiment List view. Use this option to export a single selected experiment as a delimited .txt file or a .chp file. Note that exporting to a .chp file is only possible if the source file for the experiment was an Affymetrix .chp or .cel file. Exporting to a .bed file is only available for ChIP-Seq and miRNA experiments.
Export Selected Genes
Use this option to export the currently selected genes to a tab-delimited text file format (.txt). This option is disabled if the current selection contains peak or fragment data.
Export Selected Table Rows
Use this command to export the data in the currently selected (highlighted) rows as a tab-delimited text file (.txt) or a comma separated file (.csv). This option is only available in the Gene Table, Peak Table, SNP Table, and Fragment Table views.
Export Selected Terms
This option is only available in the Gene Ontology view. Use this option to export the currently selected terms to a tab-delimited, Unicode tab delimited, or comma-separated file. For ontology terms, a “Gene Ontology ID” column will be added, which will include the ID for each term from the Gene Ontology Consortium.
Export Selected Set
This option is only available in the Set List view. Use this option to export the selected gene set to a .txt file.
Page Setup
Use this option to specify printing parameters such as which printer to use, page orientation, paper size, and paper source. Click OK to accept the changes and return to ArrayStar.
Print Preview
Use this option to view a scaled version of the current ArrayStar view as it will print. You may use the following buttons in the Print Preview window.
Use this option to print the current view. Depending upon your printer, Print may allow you to specify printing parameters such as the print range, the number of copies, whether to collate copies and whether to print the current view to a separate file.
Recent Files
Use this option to view a list of files you have recently opened, saved, or imported. This submenu is divided into three sections.
•The top section lists recently imported files; selecting a file from this section imports the file into the current project.
•The second section lists recently opened and saved projects; selecting one opens the project in place of the current one.
•The bottom section displays recently imported annotation files; selecting one from this section imports the annotation file into the current project.
Send Selection To
Provides the choice of sending selected rows of an ArrayStar table to SeqMan Pro or GenVision Pro for further analysis. For more information, see View ArrayStar Table Data in Other Lasergene Applications.
Quit
Use this option or press Ctrl + Q to close ArrayStar. If you have an unsaved project open when you use this option, you will be prompted to save.
Edit
Undo
Use this option or press Ctrl + Z to reverse the effect of the previous operation(s). If Undo is not available in a particular situation, the command will be disabled. You may select Undo multiple times to undo multiple actions.
Redo
Use this option or press Ctrl +Y to reverse the effect of the last Undo command(s). If Redo is not available in a particular situation, the command will be disabled. You may select Redo multiple times to redo multiple actions.
Cut
Use this option or press Ctrl + X to delete current the selection and place it on the clipboard to be pasted elsewhere. This option is only available in editable text fields.
Copy
Use this option or press Ctrl + C to copy the current selection to the clipboard so that it can be pasted elsewhere. In graphical views, this command copies an image of the selection; in text views, this command copies the text itself.
Paste
Use this option or press Ctrl + V to paste the contents of the clipboard into the field selected. This option is only available in editable text fields.
Clear
Use this option to delete the currently selected text.
Select All
Use this option or press Ctrl + A to select all items in the current view.
Clear Selection
Use this option to de-select the current selection.
Preferences
Use this option to open the ArrayStar Preferences dialog:
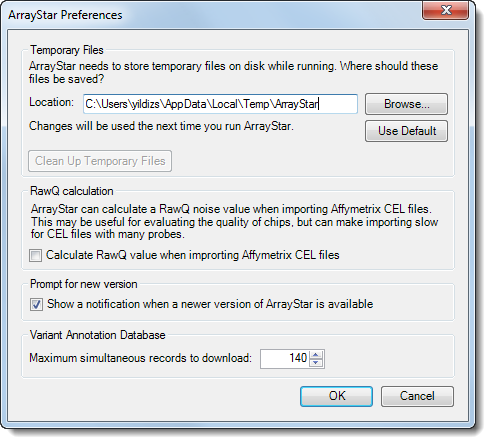
•If desired, change the location that ArrayStar uses for saving temporary files.
•Click the Browse button to locate and select a new folder.
•Click the Use Default button to return to the default directory setting: C:\Documents and Settings\<username>\Local Settings\Temp\ArrayStar
•Click Clean Up Temporary Files to delete all temporary files.
•In the RawQ calculation section, you have the option to have ArrayStar calculate a RawQ noise value when importing Affymetrix .cel files. This may be useful for evaluating the quality of chips but can make importing slow for .cel files with many probes. When calculated, the RawQ value will be displayed in the Experiment Attributes section of the Experiment List View under CHP Header Information > Summary.
•Check the box next to Calculate RawQ value when importing Affymetrix CEL files to turn on this option. By default, this preference is disabled.
•Check the box next to Show a notification when a newer version of ArrayStar is available if you want ArrayStar to automatically notify you when there is a software update.
•If using the Variant Annotation Database, input a number corresponding to the Maximum simultaneous records to download. The default value is 140 but can be changed to accommodate slower or faster computers.
Filter
Filter Selected
Use this option to perform a search on only the currently selected genes or SNPs. This option opens the Advanced Filtering window. The search criteria you enter will only be applied on those genes or SNPs that are currently selected.
Filter All
Use this option to perform a search on all of the genes or SNPs in your project. This option opens the Advanced Filtering window. The search criteria you enter will be applied on all of the genes or SNPs in the current project.
Data
Show Experiment List
Use this option to view the Experiment List for the current project.
Show Gene Table
Use this option to view the Gene Table for the current project.
Show Isoform Table
Use this option to view the Isoform Table for the current project.
Show Exon Table
Use this option to view the Exon Table for the current project.
Show Fragment Table
Use this option to view the Fragment Table for the current project.
Show Peak Table
Use this option to view the Peak Table for the current project.
Show SNP Table
Use this option to view the SNP Table for the current project.
Show Gene Ontology
Use this option to open the Gene Ontology view.
Show Set List
Use this option to view the Set List for the current project.
Show Clustering Result List
Use this option to view the Clustering Result List for the current project.
Download Annotations
Select this option to download annotation files for your data. If you are using Affymetrix data, this option will allow you to download Affymetrix Annotations. For all other data, this option will allow you to download Gene Ontology Annotations.
Delete Downloaded Annotations
Use this option to delete all of your previously downloaded annotation files. Selecting this option will display a notification listing the files to be deleted, and the amount of disk space deleting the files will free. Click Yes to delete the files listed. Click No to cancel the deletion and return to your project. This option is only enabled if you have previously downloaded annotation files.
Download Genomic Template
Use this option to download a genomic template sequence from the Download Genome Reference dialog.
Gene Information Settings
Use this option to display the Gene Information Settings window, which allows you to select the information to be displayed for each gene in the search results, Set List, and each view’s Info Pane.
Edit Data Transformations
Use this option to open the Edit Data Transformations dialog.
Exclude Genes from Project
Use this option to display the Exclude Genes window, which allows you to exclude genes from your project based on gene name prefix, by designation as a control sequence (for Affymetrix data), and/or by gene set.
Remember Selected
Use this option to save the currently selected items (e.g., genes, SNPs, exons, isoforms, etc.) as a set in your project.
Export Sequences
Use this option to export sequences for your QSeq experiments.
Clustering
Use this option to perform Hierarchical Clustering on the currently selected genes, using the default parameters.
k-Means
Use this option to perform k-Means Clustering on the currently selected genes, using the default parameters.
Advanced Clustering
Use this option to perform a clustering on the currently selected genes using customized parameters. Selecting this option displays the Advanced Clustering window.
Remember Clustering Result
Use this option to save the currently displayed clustering result. The result can then be accessed for future analysis from the Clustering Result List.
Graphs
Scatter Plot
Use this option to display the currently selected genes in the Scatter Plot view.
Heat Map
Use this option to perform Hierarchical Clustering on the currently selected genes and display the results in the Heat Map view.
Line Graph Thumbnails
Use this option to perform k-Means Clustering on the currently selected genes and display the results in the Line Graph Thumbnails view.
Venn Diagrams
Use this option to create a Venn Diagram from the currently selected genes.
Line Graph
Use this option to create a Line Graph from the currently selected genes.
Set Colors
Use this option to display the Set Colors window for the current view, which allows you customize the color settings to suit your preferences.
Set Styles
Use this option to customize the display settings for the current view:
Statistics
Fisher's Exact Test Signal Search
Use this option with non-Variants projects to perform Fisher's Exact Test. See Fisher's Exact Test Signal/SNP Search for more information.
Fisher's Exact Test SNP Search
Use this option with Variants projects to perform Fisher's Exact Test in Variants projects. See Fisher's Exact Test Signal/SNP Search for more information.
Odds Ratio SNP Search
Use this option with Variants projects to perform the Odds Ratio test in Variants projects. See Odds Ratio SNP Search for more information.
Standard deviation
Use this option to calculate the Standard Deviation for the currently selected experiment. Selecting this command will display the Statistics Parameters window. Adjust the parameters as necessary, and then click Calculate. The resulting values will be added to the Gene Table view.
Coefficient of Variation
Use this option to calculate the Coefficient of Variation for the currently selected experiment. Selecting this command will display the Statistics Parameters window. Adjust the parameters as necessary, and then click Calculate. The resulting values will be added to the Gene Table view.
Variance
Use this option to calculate Variance for the currently selected experiment. Selecting this command will display the Statistics Parameters window. Adjust the parameters as necessary, and then click Calculate. The resulting values will be added to the Gene Table view.
Cross-R2
Use this command to perform an R2 statistical test for all pairwise combinations of selected experiments in the project and to produce a table of the results. This command allows you to perform batch comparisons of replicates and samples.
Validation Control Accuracy
Use this option to generate a Validation Control Accuracy report for certain types of SeqMan NGen assemblies. See Validation Control Accuracy for more information.
Student’s t-test
Use this option to apply the Student’s t-test to the currently selected experiments. This command is only enabled for QSeq and Microarray experiments. Selecting this command will display the Statistics Parameters window. Adjust the parameters as necessary, and then click Calculate. The resulting values will be added to the Gene Table view. The Student's t-test can also be accessed using the drop-down menu in the Scatter Plot info pane.
Moderated t-test
Use this option to apply the Moderated t-test to the currently selected experiments. This command is only enabled for QSeq and Microarray experiments. Selecting this command will display the Statistics Parameters window. Adjust the parameters as necessary, and then click Calculate. The resulting values will be added to the Gene Table view. The Moderated t-test can also be accessed using the drop-down menu in the Scatter Plot info pane.
Use this option to apply the F-Test (ANOVA) to the currently selected experiments. Selecting this command will display the Statistics Parameters window. Adjust the parameters as necessary, and then click Calculate. The resulting values will be added to the Gene Table view.
View
Duplicate View
Use this option to create a copy of the current view. This option is only enabled if one of the following views is active:
Close View
Use this option or press Ctrl + W to close the current view. You may also close a view by right-clicking on the view’s title bar and selecting Close.
Dock/Undock View
Use this option to dock or undock the current view. If the current view is docked, this option will undock the view. If the current view is undocked, this option will re-dock the view. Views may also be docked/undocked by right-clicking in the view’s title bar and selecting Dock or Undock.
Edit View Name/Notes
Use this option to edit the title and notes for the current view. You can also edit the view’s title and notes by right-clicking on the view’s title bar and selecting Edit View Name/Notes.
[list of open views]
At the bottom of the View menu, a list of all open views is displayed. Select a view from the list to bring it to the front and make it the active view
Help
ArrayStar Help
Use this option or press F1 to open the ArrayStar Help file. ArrayStar will automatically direct you to the Help topic for the current view, however you can navigate to or search all of the available topics.
ArrayStar Tutorials
Use this option to be directed to the DNASTAR website to download ArrayStar tutorials.
Software Updates
Use this option to be directed to the DNASTAR website for software updates. You must have a valid User ID and Subscriber Number to download software.
Technical Support Request
Use this option to be directed to the DNASTAR website to request technical support.
About ArrayStar
Use this option to view the registration and version information of this copy of ArrayStar.