Note: This topic is not applicable to BAM-based projects.
To access the Search dialog, select Edit > Find.
Alternatively, click on the search box found near the bottom left corner of the Alignment View, the Contig Info Report, the Show Seq Info window, or the Project Report. This box will either contains the words Unspecified Search, or your last specified search criteria.

The Search dialog is displayed:
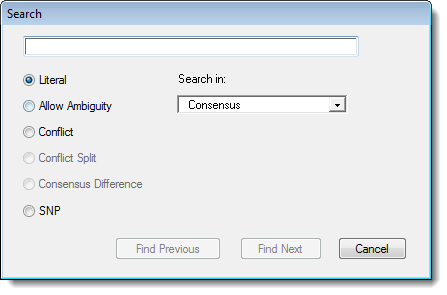
Select the type of search you wish to perform. (Note that not all options are available in every view.)
•Literal - Searches for perfect matches only. You can use IUB ambiguity codes in literal sequence searches, but these will match only the same ambiguity code in the sequence. For example, searching for “S”—the ambiguity code for G or C—will locate only S, and neither G nor C. This can be useful for locating specific heterozygotes. To search for gaps, enter a dash (-) into the text box.
•Allow Ambiguity - recognizes IUB ambiguity codes in the query. If you enter the query GATS, for example, you would find GATC, GATG and GATS.
•Conflict - locates any base that differs from one or more bases in other sequences in the same column of the alignment. SeqMan Pro will find and highlight single conflicting residue columns or consecutive groups of conflicting columns.
•Conflict Split - locates, one at a time, the conflict patterns which would be found by selecting the Suggest Conflict Splits command.
•Consensus Difference - locates differences between the two active consensus sequences, as displayed at the top of the Alignment View.
•SNP - locates bases that have been identified as putative, confirmed, or rejected variants.
Note: Conflict Split, Consensus Difference, and SNP searches may only be performed for the consensus.
For literal or ambiguous searches, enter a text string in the field at the top of the dialog.
If you accessed the Search dialog from the Alignment View, specify where you want SeqMan Pro to search by using the Search in list:
•Choose Consensus to search only for items in the consensus sequence.
•Choose Selected Sequence to search only the sequence where the cursor is currently located.
•Choose All Sequences in Contig to search in each constituent sequence, but not in the contig itself.
Click Find Previous or Find Next to perform a search. After initiating a search, you can click the forward ( > > ) or reverse (<<) arrowheads in the bottom left corner of the window to jump to the next or previous find. After reaching either end of a sequence, SeqMan Pro wraps around and continues searching from the other end.