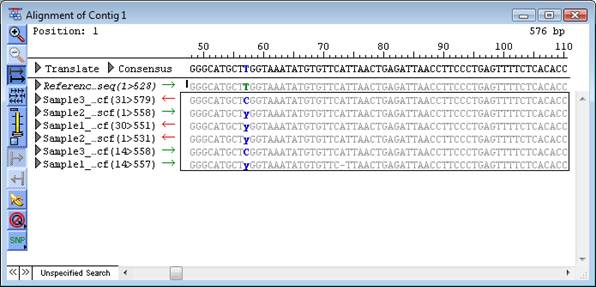
To display variants in the Alignment View, select Variant > Show Variants.
The Alignment View changes as follows:

•If the reference sequence is from a GenBank file, bases annotated as features of type “variation” are displayed in green, by default.
•Bases in sample sequences are also colored to distinguish non-variants and types of variants throughout the view. These colors can be changed to suit your preferences from the Variant Discovery parameters. Default colors are:
o Blue for putative variants
o Green for confirmed variants. Note that all reference variants are colored as confirmed variants.
o Red for rejected variants
o Gray for non-variants
•A base in a sample sequence that SeqMan Pro has determined is a heterozygous putative variant is displayed using the appropriate ambiguity code.
•To hide variants in the Alignment View, select Variant > Hide Variants to return to the normal display.
Note: You cannot edit when variants are displayed; you can only edit in the normal display.