Option
Description
Table
1000Gp3_GF
Gene frequency for a given population, from the 1000 Genomes Project Phase3 via dbNSFP Version 2.7. Phase3 comprises genomic data from twenty-six populations distributed among five super populations.
In the Manage Columns dialog (Using the Manage Columns Dialog), the main category at the top of the tree displays alternative gene frequencies for all populations combined. Below this is a list of super populations, any of which can be expanded to reveal sub-populations.
|
Super Population |
Sub-Population |
|
AFR - African |
ACB - African Caribbeans in Barbados |
|
ASW - Americans of African Ancestry in SW USA | |
|
ESN - Esan in Nigeria | |
|
GWD - Gambian in Western Divisions in The Gambia | |
|
LWK - Luhya in Webuye, Kenya | |
|
MSL - Mende in Sierra Leone | |
|
YRI - Yoruba in Ibadan, Nigeria | |
|
AMR - Ad Mixed American |
CLM - Colombians from Medellin, Colombia |
|
MXL - Mexican Ancestry from Los Angeles USA | |
|
PEL - Peruvians from Lima, Peru | |
|
PUR - Puerto Ricans from Puerto Rico | |
|
EAS - East Asian (equivalent to ASN in the 1000 Genomes Catalog) |
CDX - Chinese Dai in Xishuangbanna, China |
|
CHB - Han Chinese in Bejing, China | |
|
CHS - Southern Han Chinese | |
|
JPT - Japanese in Tokyo, Japan | |
|
KHV - Kinh in Ho Chi Minh City, Vietnam | |
|
EUR - European |
CEU - Utah Residents (CEPH) with Northern and Western European ancestry |
|
FIN - Finnish in Finland | |
|
GBR - British in England and Scotland | |
|
IBS - Iberian population in Spain | |
|
TSI - Toscani in Italia | |
|
SAS - South Asian |
BEB - Bengali from Bangladesh |
|
GIH - Gujarati Indian from Houston, Texas | |
|
ITU - Indian Telugu from the UK | |
|
PJL - Punjabi from Lahore, Pakistan | |
|
STU - Sri Lankan Tamil from the UK |
S1
1000Gp3_MAF…
Alternative allele frequency for a given population, from the 1000 Genomes Project Phase3 via dbNSFP Version 2.7. Phase3 comprises genomic data from twenty-six populations distributed among five super populations.
In the Manage Columns dialog (Using the Manage Columns Dialog), the main category at the top of the tree displays alternative allele frequencies for all populations combined. Below this is a list of super populations, any of which can be expanded to reveal sub-populations. See the table above for descriptions.
S1
/[annotation type]
Available GenBank-style annotations vary based on the annotations you have imported. If the data in your project was imported through the Data Import Wizard, any columns you designated as “Description” during import will appear in this list.
Accessible annotations also vary depending on the table from which you accessed the Manage Columns dialog. Each of the tables whose abbreviation appears on the right offers only a subset of all available annotations.
E, G, I
/qseq_name
Unambiguous name assigned by QSeq to avoid duplicate names in a genome. When duplicate names are found, a number is added to disambiguate.
G20, I20
aaAlt
The alternative amino acid. A period is shown if the variant is a splicing site SNP (i.e., 2 base pairs on each end of an intron).
S1
aaPos
The amino acid position in relation to the protein. A negative one (-1) appears if the variant is a splicing site SNP (i.e., 2 base pairs on each end of an intron).
S1
aaRef
The reference amino acid. A period is shown if the variant is a splicing site SNP (i.e., 2 base pairs on each end of an intron).
S1
adjusted P value
The FDR/Benjamini-Hochberg multiple testing correction value, also known as the “Q value.” This column is available if Bioconductor’s DESeq2 was used as the normalization method, and also applies to T tests and F tests when the FDR/Benjamini-Hochberg multiple testing correction is applied (as it is by default).
G25, I25
amino acid change
The change(s) in the protein sequence using the nomenclature and conventions established by the Human Genome Variation Society (HGVS). The cell text ‘p.(=)’ signifies a synonymous change, while an empty cell denotes a non-coding region.
S
aspect
See the GO Annotation File Format 2.0 Guide for a description.
G2
assigned_by
See the GO Annotation File Format 2.0 Guide for a description.
G2
base mean
The mean of normalized counts for that gene across all samples.
G25, I25
binding protein
The binding protein used for the experiment. Binding proteins are assigned to each experiment by you in the Create Binding Proteins step of the Project Setup Wizard.
F3, P3
called seq
The base or sequence of the permutation. This might be a gap ‘-‘ for a deletion, multiple bases for an insertion, or the same as Ref Seq for a non-change. Multiple called bases are shown with dividers, e.g. T|C, A|-, etc. An empty cell denotes no Database of Single Nucleotide Polymorphism (dbSNP) information, while (NC) refers to “no coverage.” This data derives from the SNP Sequence column in the Data Import Wizard.
S
CCDS_id
CCDS ID, from the HUGO Gene Nomenclature Committee (HGNC).
G4
chr
Chromosome number, from the HUGO Gene Nomenclature Committee (HGNC).
G4
classification
The category and type of permutation (coding, synonymous, frameshift, etc.)
S
clinvar_clnsig
The clinical/pathological significance from the ClinVar database.
•Benign (2)
•Likely benign (3)
•Likely pathogenic (4)
•Pathogenic (5)
•Drug response (6)
•Histocompatibility (7)
S1
clinvar_rs
The reference SNP (rs) number from the ClinVar database.
S1
clinvar_trait
The trait identifier (e.g., CUI, HPO, etc.) from the ClinVar database.
S1
codonpos
The position on the codon (1, 2 or 3).
S1
combined SNP seq6
The raw coalesced/combined SNP sequence as recorded by SeqMan NGen.
S5
contig pos
The position on a gapped contig corresponding to this chromosome (for SeqMan and SeqMan NGen data only).
S
cooks
The Cook’s distance for that gene. It is a measure of how much a single sample is influencing the fitted coefficients for that gene. Large values indicate an outlier count.
G25, I25
COSMIC ID
Catalogue of Somatic Mutations in Cancer (COSMIC) ID with hyperlink to the COSMIC entry.
S5
date
See the GO Annotation File Format 2.0 Guide for a description.
G2
DB
See the GO Annotation File Format 2.0 Guide for a description.
G2
db:Reference
See the GO Annotation File Format 2.0 Guide for a description.
G2
db_Object _ID7
See the GO Annotation File Format 2.0 Guide for a description.
G2
db_Object_Name7
See the GO Annotation File Format 2.0 Guide for a description.
G2
db_Object_Symbol7
See the GO Annotation File Format 2.0 Guide for a description.
G2
db_Object_Synonym7
See the GO Annotation File Format 2.0 Guide for a description.
G2
DB_Object_Type
See the GO Annotation File Format 2.0 Guide for a description.
G2
dbSNP ID
The Database of Single Nucleotide Polymorphism (dbSNP) ID with hyperlink to the dbSNP entry.
S5
depth
The depth of coverage for the SNP.
S8
detected exons
The number of exons detected in the gene
G9
disease_description
Disease(s) the gene causes or with which it is associated, from Uniprot.
G4
DNA Change
The change(s) in the DNA sequence using the nomenclature established by the Human Genome Variation Society (HGVS).
S
downstream gene
The name of the nearest downstream gene within 100 kilobases of the fragment or peak. If there is an intersecting gene, no downstream gene will be listed.
F, P
end
The end position of the fragment along the template sequence.
F, P
Ensembl_gene
Ensembl gene ID, from the HUGO Gene Nomenclature Committee (HGNC).
G4
Entrez_gene_id
Entrez gene ID, from the HUGO Gene Nomenclature Committee (HGNC).
G4
ESP6500_MAF
Alternative allele frequency for a given population, from the NHLBI Exome Sequencing Project’s ESP6500 data set. The main category at the top of the tree displays alternative allele frequencies for all populations combined. Below this are the two super populations:
•AA – African American population only.
•EA – European American population only.
S1
essential_gene
Essential I or Non-essential (N) phenotype-changing based on Mouse Genome Informatics (MGI) database (Georgi et al., 2013|topic=Research References).
G4
evidence code
See the GO Annotation File Format 2.0 Guide for a description.
G2
experiment ID
The name of the experiment for the peak, as shown in the Experiment List.
P
expression (egenetics)
Tissues/organs in which the gene is expressed, from egenetics data from BioMart.
G4
Expression(GNF/Atlas)
Tissues/organs in which the gene is expressed, from GNF/Atlas data from BioMart.
G4
FDR
The False Discovery Rate (FDR) represents the likelihood that the peak is not valid. The FDR signal value is only available if control data are present and MACS Peak Detection is used for peak discovery.
F
feature type
The type of feature as annotated in the template sequence for the gene (e.g. CDS, attenuator, C_region, etc.). The genes for the template are defined during Set Up Preprocessing.
The annotations available in the drop-down list will vary based on the annotations you have imported. If the data in your project was imported through the Data Import Wizard, any columns you designated as Description during import will appear in this list.
E12, G12, I12
fold enrichment
This column is available only in the Peak Table for ChIP-Seq experiments and is available through the Add/Manage Columns under IP Peak Values. Fold Enrichment is calculated from the enriched tags in that peak region and the local lambda of Poisson distribution from the nearby regions. Note that it is not the same analysis used for the MFOLD value. Many reported peaks may have Fold Enrichment values less than the MFOLD cutoff.
P
fragment ID
The ID for the fragment of the genome along which the peak occurs. See the Fragment Table or more information.
G11, P
frameshift14
This change is caused by indels that are not evenly divisible by three.
G13
full alleles6
The set of raw alleles as recorded in the VCF file, including the flanking 5’ anchor base that is considered extraneous by ArrayStar.
S5
full ref seq6
The reference sequence of the raw coalesced/combined SNP sequence as recorded by SeqMan NGen.
S5
function_description
Function description of the gene from Uniprot.
G4
gene_full_name
Gene full name from the HUGO Gene Nomenclature Committee (HGNC).
G4, S4
gene_name, gene name
Gene symbol (e.g. recA, thrL, etc.), from the HUGO Gene Nomenclature Committee (HGNC).
G4, S4
gene_old_names
Old gene symbol, from the HUGO Gene Nomenclature Committee (HGNC).
G4
gene_other_names
Other gene names, from the HUGO Gene Nomenclature Committee (HGNC).
G4
genotype
Whether this SNP call indicates the reference or not, and (for diploids) whether this calls a homozygous or heterozygous genotype.
S
GERP Score
Genomic Evolutionary Rate Profiling (GERP) score.
S5
GERP++_NR
Genomic Evolutionary Rate Profiling (GERP++10) neutral rate (NR) – the number of substitutions expected under conditions of neutrality.
S1
GERP++_RS
Genomic Evolutionary Rate Profiling (GERP++10) rejected substitutions (RS) – the number of substitutions expected under neutrality minus the number of substitutions ‘‘observed’’ at the position. Scores range from 1 to 6.18.The larger the score, the more conserved the site.
S1
GERP++_RS_rankscore
Genomic Evolutionary Rate Profiling (GERP++10) RS scores were ranked among all GERP++ RS scores in dbNSFP. The “rankscore” is the ratio of the rank of the score over the total number of GERP++ RS scores in dbNSFP.
S1
GO ID
See the GO Annotation File Format 2.0 Guide for a description.
G2
GO_Slim_biological_process
Terms for biological process from Gene Ontology (GO) Slim.
G4
GO_Slim_cellular_component
Terms for cellular component from Gene Ontology (GO) Slim.
G4
GO_Slim_molecular_function
Terms for molecular function from Gene Ontology (GO) Slim.
G4
highest exon
The largest copy number value of any discovered exon in the gene. For RPKM-CN or zRPKM-normalized workflows, exons are ignored if they are below 1 RPKM. For zRPKM workflows, exons are also ignored if there is a systematic lack of depth across the same exon in all experiments.
G9
inframe indel14
An insertion or deletion within a coding region whose length is divisible by 3.
•For insertions, the type is followed by the word Conservative when inserted bases occur between two codons, and by the word Disruptive when the inserted bases occur within a codon.
•For deletions, the type is followed by the word Conservative when deleted bases begin at the first position of a codon and end at the last position of a codon, and by the word Disruptive when the deleted bases start at position 2 or 3 of a codon and end at position 1 or 2, respectively, of another codon.
G13
Interactions (BioGRID)
Other genes with which the gene interacts, from BioGRID. The gene name is followed by the PubMed ID in square brackets.
G4
Interactions (ConsensusPathDB)
Other genes with which the gene interacts, from ConsensusPathDB. The gene name is followed by the PubMed ID in square brackets.
G4
Interactions (IntAct):
Other genes with which the gene interacts, from IntAct. The gene name is followed by the PubMed ID in square brackets.
G4
Interpro_domain
The domain or conserved site on which the variant is located. Domain annotations come from Interpro database. The number in the brackets following a specific domain is the count of times Interpro assigns the variant position to that domain, typically coming from different predicting databases.
S1
intersecting genes
The names of any genes in the template that intersect the fragment or peak. The genes for the template are defined during Set Up Preprocessing.
F, P
known_rec_info
Known recessive status of the gene (MacArthur et al., 2012).
•lof-tolerant – seen in homozygous state in at least one 1000G individual.
•recessive – known OMIM recessive disease.
G4
lfc SE
The standard error estimate for the log2 fold change estimate.
G25, I25
length
The length of the fragment, equal to the end position minus the start position.
F, P
linear expression level
Though the log2 format is displayed by default, the option for displaying linear values is available in some workflows (see Using the Manage Columns Dialog). The linear signal is equal to (total_)raw_count + repeat_distrib_count, with any selected normalization applied.
linear rlog reads
The rlog values for each gene transformed back into the linear scale. The linear rlog value for the control set is also displayed. This value differs from raw DESeq2 in that it also includes any user-applied data transformations.
G25, I25
log2 expression level
The log2 signal columns in the Gene Table, Fragment Table, Peak Table, Exon Table and Isoform Table can display positive or negative values:
•Positive – the underlying value is greater than one. A value of +1 indicates a two-fold increase. If you have loaded data which comprise a ratio, a positive value in this column denotes up-regulated gene expression.
•Negative – the underlying value is less than one. A value of -1 indicates a two-fold decrease. If you have loaded data which comprise a ratio, a negative value in this column denotes down-regulated gene expression.
Some examples of log2 equivalents are shown below:
|
Linear Value |
Log2 Equivalent |
|
1 |
0 |
|
0.25 |
-2 |
|
64 |
6 |
In addition to displaying this column, you can also over the mouse over any signal column to display a tool tip showing both rounded and unrounded log2 values.
log2 fold change
The effect size estimate indicating how much the gene expression seems to have changed in the test sample relative to control. This value is reported on a logarithmic scale to base 2.
G25, I25
lowest exon
The smallest copy number value of any discovered exon in the gene. For RPKM-CN or zRPKM-normalized workflows, exons are ignored if they are below 1 RPKM. For zRPKM workflows, exons are also ignored if there is a systematic lack of depth across the same exon in all experiments.
G9
LRT_converted_rankscore15
Likelihood Ratio Test (LRT) scores were first converted as LRTnew = (1 – LRTori * 0.5) if Omega < 1, or LRTnew = (LRTori * 0.5) if Omega ≥ 1. Then LRTnew scores were ranked among all LRTnew scores in dbNSFP. The rankscore is the ratio of the rank over the total number of the scores in dbNSFP. The scores range from 0.00166 to 0.85682.
S1
LRT_pred15
Likelihood Ratio Test (LRT) predictions are based on several criteria including the LRT score.
•D(eleterious) – non-synonymous SNP (NS) needs to fulfill three requirements: 1) from a codon defined by LRT as significantly constrained (LRTori <0.001 and ω <1); 2) from a site with >10 eutherian mammals’ alignments; 3) the alternative AA is not presented in any of the eutherian mammals
•N(eutral) – NS needs to fulfill either of the two requirements: (i) the alternative AA is presented in at least one of the eutherian mammals, or (ii) from a codon defined by LRT as not significantly constrained (LRTori >0.001 or ω >1) and with >10 eutherian mammals’ alignments
•U(nknown) – Alternative amino acid from alignment positions with <10 eutherian mammals.
S1
LRT_score15
The p-value from the Likelihood Ratio Test (LRT). The smaller the score the more likely the SNP has a damaging effect. Scores range from 0 to 1.
S1
mean values from DESeq2
This value is used for DNASTAR diagnostic use only.
G25, I25
MIM_disease
MIM disease name(s) with MIM ID(s) in square brackets from Uniprot.
G4
MIM_id
MIM gene ID, from the HUGO Gene Nomenclature Committee (HGNC).
G4
MIM_phenotype_id
MIM ID(s) of the phenotype the gene causes or with which it is associated, from Uniprot.
G4
MM Count 1
Count of articles in the Mastermind Genomic Search Engine with cDNA matches for this specific variant.
S1
MM Count 2
Count of articles in the Mastermind Genomic Search Engine with variants either explicitly matching at the cDNA level or given only at protein level.
S1
MM Count 3
Count of Mastermind articles in the Mastermind Genomic Search Engine, including other DNA-level variants resulting in the same amino acid change.
S1
MM Gene
Genes for this variant from the Mastermind Genomic Search Engine.
S1
MM HGVS
HGVS genomic notation for this variant from the Mastermind Genomic Search Engine.
S1
MM ID 3
Variant identifiers in the Mastermind Genomic Search Engine, as gene:key, for MMCNT3.
S1
MM URI 3
Search URI for articles in the Mastermind Genomic Search Engine, including other DNA-level variants resulting in the same amino acid change.
S1
mu
Mean fitted count values from individual replicates.
G25, I25
MutationTaster_converted_rankscore16
MutationTaster MTori scores were first converted. If the prediction is “A” or “D,” then Mtnew = MTori; if the prediction is “N” or “P”, then Mtnew = (1-Mtori). Mtnew scores were then ranked among all Mtnew scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of Mtnew scores in dbNSFP. Scores range from 0.0931 to 0.80722.
S1
MutationTaster_pred16
MutationTaster prediction.
|
Designation |
Score |
Description |
|
Disease_causing_(A)utomatic |
|
Known to be deleterious; variant is marked as probable-pathogenic or pathogenic in ClinVar |
|
(D)isease_causing |
> 0.5 |
Probably deleterious |
|
(N) – Polymorphism |
< 0.5 |
Probably harmless |
|
(P)olymorphism_automatic |
|
Known to be harmless; each of the three possible genotypes is observed in the 1000 Genomes Project |
S1
MutationTaster_score16
The MutationTaster probability that prediction is correct. Scores range from 0 to 1. The larger the score the more likely it is correct.
S1
nonsense14
This change results in a premature stop codon and a truncated, incomplete, and usually nonfunctional protein product.
G13
non-synonymous14
This change alters the amino acid sequence coded for in this (translated) coding region. This category includes Nonsense, Frameshift, No-start and No-stop changes. To derive the number of simple substitutions, display all of the options and subtract the four listed changes from the Non-synonymous count.
G13
no-start14
Change resulting in the absence of a start codon.
G13
no-stop14
Change resulting in the absence of a stop codon.
G13
notes
User-created notes about the gene.
G
P not ref
The probability that this position does not match the reference. For combined SNPs and indels, P not ref will be the minimum of the P not refs in the used columns.
S8
P(HI)
Estimated probability of haploinsufficiency of the gene (Huang N et al., 2010|topic=Research References).
G4
P(rec)
Estimated probability that gene is a recessive disease gene (MacArthur et al., 2012|topic=Research References).
G4
pathway(ConsensusPathDB)
Pathway(s) to which the gene belongs, from ConsensusPathDB.
G4
pathway(Uniprot)
Pathway(s) to which the gene belongs, from Uniprot.
G4
peak ID
The IDs of any peaks that are present within the fragment. See the Peak Table for more information.
F, G11
phastCons100way_vertebrate17
The PhastCons conservation score, based on the multiple alignments of 100 vertebrate genomes (including human) and a phylo-HMM. The larger the score, the more conserved the site. Scores ranges from 0 to 1.
S1
phastCons100way_vertebrate_rankscore17
PhastCons100way_vertebrate scores (PhastCons) were ranked among all phastCons100way_vertebrate scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of phastCons100way_vertebrate scores in dbNSFP.
S1
phastCons46way_placental17
The PhastCons conservation score, based on the multiple alignments of 33 placental mammal genomes (including human) and a phylo-HMM. The larger the score, the more conserved the site. Scores ranges from 0 to 1.
S1
phastCons46way_placental_rankscore17
PhastCons46way_placental scores (PhastCons)were ranked among all phastCons46way_placental scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of phastCons46way_placental scores in dbNSFP.
S1
phastCons46way_primate17
The PhastCons conservation score, based on the multiple alignments of 10 primate genomes (including human) and a phylo-HMM. The larger the score, the more conserved the site. Scores ranges from 0 to 1.
S1
phastCons46way_primate_rankscore17
PhastCons46way_primate scores (PhastCons) were ranked among all phastCons46way_primate scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of phastCons46way_primate scores in dbNSFP.
S1
phyloP100way_vertebrate18
The phylogenetic p-values (phyloP) conservation score, representing the –log p-value under the null hypothesis of neutral evolution at the site based on the multiple alignments of 100 vertebrate genomes (including human). The larger the score, the more conserved the site. Scores ranges from -20 to 10.
S1
phyloP100way_vertebrate_rankscore18
PhyloP100way_vertebrate scores (phyloP) were ranked among all phyloP100way_vertebrate scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of phyloP100way_vertebrate scores in dbNSFP.
S1
phyloP46way_placental18
The phylogenetic p-values (phyloP) conservation score, which represents the –log p-value under the null hypothesis of neutral evolution at the site based on the multiple alignments of 33 placental mammal genomes (including human). The larger the score, the more conserved the site. Scores range from –11.958 to 3.
S1
phyloP46way_placental_rankscore18
PhyloP46way_placental scores (phyloP) were ranked among all phyloP46way_placental scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of phyloP46way_placental scores in dbNSFP.
S1
phyloP46way_primate18
The phylogenetic p-values (phyloP) conservation score, which represents the –log p-value under the null hypothesis of neutral evolution at the site based on the multiple alignments of 10 primate genomes (including human). The larger the score, the more conserved the site. Scores ranges from -8.176 to 0.66.
S1
phyloP46way_primate_rankscore18
PhyloP46way_primate scores (phyloP) were ranked among all phyloP46way_primate scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of phyloP46way_primate scores in dbNSFP.
S1
Pinnacle
The position on the template sequence where the peak reaches its top height.
P
pValue
For QSeq Peak Finder and MACS Peak Detection results, pValue score for each peak which is equal to the log10 likelihood that the peak is “valid” multiplied by -10.
F
Q call
The Phred-like quality score of the called genotype. It is a measure of the confidence that the SNP is present in the sample on a 0-60 log10 scale. For combined SNPs and indels, Q call will be the minimum of all available columns at that reference position.
S8
QSeq ID
QSeq-generated ID used to link data between tables and to link workflows together.
G20, E20, I20
qualifier
See the GO Annotation File Format 2.0 Guide for a description.
G2
raw_count19
The number of reads that were initially and uniquely assigned to an isoform, fragment or peak (i.e., reads placed only once). The raw_count value is not normalized, and repeated reads are excluded. It remains constant regardless of any normalization applied to the experiment
If there are duplicated genes, none of the reads will map uniquely. This makes it possible for a read to have a raw count of zero, but still have non-zero signal and fold change values. See example at the very bottom of this topic.
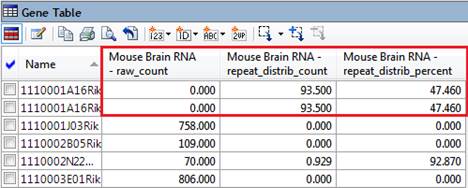
Note that the “Uniquely mapped count” is the total number of unique aligned reads with no normalization applied. This value is used in the calculation of (total_)raw_count, but not explicitly displayed in ArrayStar.
E, F, I21
raw DESeq2,
raw DESeq2-Gene
The rlog values from individual replicates.
G25, I25
raw P value
The standard non-FDR corrected P value. This column is only available in the Gene and Isoform tables if you choose a Bioconductor statistic (DESeq2, DESeq2-Local, edgeR, or edgeR-Local) in the Set Up Preprocessing page. In cases where the counts for a gene were zero or contained an extreme outlier(s), DESeq2 will return a value of NA (“not available”). ArrayStar displays these values as “-----”.
G25, I25
raw_repeat_count
The total fraction of repeat reads that map to the peak. For example, if one read maps equally well to two peaks, each peak would have a raw_repeat_count of 0.5. If multiple repeat reads map to a single peak, the proportions are summed to get the raw_repeat_count. This column is only available for QSeq Peak Finder results when repeat handling is used.
F, I21
read_count
The total number of reads.
I25
ref seq
Reference sequence at the same position as the SNP.
S
ref ID
The base at this position on the reference chromosome.
S
ref pos
The position on the reference chromosome. This data derives from the SNP Position column in the Data Import Wizard.
S
refcodon
The reference codon.
S1
Refseq_id
Refseq gene ID, from the HUGO Gene Nomenclature Committee (HGNC).
G4
Regularized logarithm (rlog) values from Bioconductor
Same as linear rlog reads.
G25, I25
repeat_distrib_count19
The proportional number of repeated reads assigned to this exon, gene or isoform. This number remains constant regardless of any normalization applied to the experiment. See example at the very bottom of this topic.
E21, I21
repeat_distrib_percent19
A rough estimate of the percentage of the total repeated reads which could have been assigned and that were assigned. In other words, he percentage of repeat_distrib_count in raw_repeat_count. This number remains constant regardless of any normalization applied to the experiment. See example at the very bottom of this topic.
E21, G21, I21
rlog
Same as linear rlog reads.
G25, I25
RPKM
The RPKM calculated for each exon (reads per million mapped reads in the experiment per kilobase of the exon). This option is only available if you chose RPKM-CN or zRPKM normalization.
E
RPKM median
The median RPKM of this exon in all applicable experiments (excluding RPKM < 1). This option is only available if you chose zRPKM normalization.
E
RPKM std dev
The standard deviation of RPKM of this exon in all applicable experiments (excluding RPKM < 1). This option is only available if you chose zRPKM normalization.
E
rs_dbSNP141
The reference SNP (rs) number from dbSNP 141.
S1
SIFT_converted_rankscore22
To obtain the rankscore, Sorting Intolerant from Tolerant (SIFT) scores were first converted to SIFTnew = (1-SIFTori), then ranked among all SIFTnew scores in dbNSFP. The rankscore is the ratio of the rank the SIFTnew score over the total number of SIFTnew scores in dbNSFP. If there are multiple scores, only the largest (most damaging) rankscore is presented. Rankscores range from 0.02654 to 0.87932.
|
Designation |
Rankscore |
|
(D)amaging |
> 0.55 |
|
(T)olerated |
|
S1
SIFT_pred22
Sorting Intolerant from Tolerant (SIFT) prediction is based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences.
S1
SIFT_score22
Sorting Intolerant from Tolerant (SIFT) score is based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences. Scores range from 0 to 1. The smaller the score the more likely the SNP has a damaging effect.
|
Designation |
SIFTori |
|
(D)amaging |
< 0.05 |
|
(T)olerated |
> 0.05 |
S1
SiPhy_29way_logOdds23
The SiPhy score based on 29 mammals genomes. The larger the score, the more conserved the site. Scores ranges from 0 to 37.9718.
S1
SiPhy_29way_logOdds_rankscore23
SiPhy_29way_logOdds scores (SiPhy) were ranked among all SiPhy_29way_logOdds scores in dbNSFP. The rankscore is the ratio of the rank of the score over the total number of SiPhy_29way_logOdds scores in dbNSFP.
S1
SiPhy_29way_pi23
The estimated stationary distribution of A, C, G and T at the site, using a SiPhy algorithm that includes biased substitution patterns based on 29 mammals genomes.
S1
SNP Base6
For diagnostic purposes only.
S5
SNP Gene Name
If the data in your project were imported through the Data Import Wizard, any columns you designated as “Gene IDs” will appear in this field.
G13
SNP%
The percentage of the sequence at this position in the assembly which varied from the reference.
S
SNPs14
A single-nucleotide polymorphism. This category is a summation of SNPs from each individual category (e.g. Nonsense, No-stop, etc.). Only substitutions are included in the count; indels are excluded.
G13
source file24
The name and location of the template sequence file, or the template sequence on which the fragment occurs. If you downloaded templates from NCBI during Set Up Preprocessing, this will be the accession number for the template. If this column is used in a table, you can hover over the sequence name in the table for more information.
E11, F11, G11, I11, P11
source seq length
The length, in base pairs, of the template sequence.
E11, F11, G11, I11, P11
source sequence
The template sequence on which the gene occurs. If you downloaded templates from NCBI during Set Up Preprocessing, this will be the accession number for the template. If this column is used in a table, you can hover over the sequence name in the table for more information.
E11, F11, G11, I11, P11
splice14
This change includes all SNPs that modify splicing. This category is a count of flags which can apply to any of the other categories.
G13
start
The start position of the fragment along the template sequence.
F, P
synonymous14
This change does not alter the amino acid sequence coded for in this (translated) coding region.
G13
target length
The target length of the fragment, equal to the end position minus the start position.
E11, G11, I11
target range
E11, G11, I11
taxon
See the GO Annotation File Format 2.0 Guide for a description.
G2
top height
The depth of the peak, in reads, at its highest point.
P
total RPKM
Total reads assigned per kilobase of target per million mapped reads.
G9
total_raw_count
This value can be applied to the Gene Table. It is similar to raw_count, available in the Isoform Table, except that it is totaled across the isoforms for that gene.
G9, G25, I25
total_read_count
This value can be applied to the Gene Table. It is similar to read_count, available in the Isoform Table, except that it is totaled across the isoforms for that gene.
G25
total_repeat_distrib_count
This value can be applied to the Gene Table. It is similar to repeat_distrib_count, available in the Isoform Table, except that it is totaled across the isoforms for that gene.
G9, G25
trait_association (GWAS)
Trait(s) with which the gene is associated, from the Genome-Wide Association Studies (GWAS) catalog.
G4
type
Whether it is a literal SNP or an indel. Available for SeqMan Pro data only.
S
ucsc_id
UCSC gene ID, from the HUGO Gene Nomenclature Committee (HGNC).
G4
Uniprot_aapos
Uniprot amino acid position.
S1
Uniprot_acc
Uniprot accession number.
G4, S1
Uniprot_id
Uniprot ID number.
G4, S1
Upstream Gene
The name of the nearest upstream gene within 100 kilobases of the fragment or peak. If there is an intersecting gene, no upstream gene will be listed.
F, P
user ID
The position ID from a VCF SNP table embedded into an .assembly project that was imported to ArrayStar. This column type is not available if you imported the VCF SNP data into ArrayStar separately.
S5
vst
Variance Stabilizing Transformation value in log2 scale.
G25, I25
Wald stat
Value from the Wald test which divides the log2 fold change by the standard error to calculate a P value compared to a normal distribution.
G25, I25
with (or) from
See the GO Annotation File Format 2.0 Guide for a description.
G2