Viewing and working with variants in a graphical display:
- SeqMan Pro – Variants are displayed as colored nucleotides in the Alignment view, and can be spotted more easily using Variant > Show Variants.
- SeqMan Ultra – Variants can be displayed in the Alignment view using a variety of foreground and background color schemes. These displays are customized using the Style panel.
Unlike SeqMan Pro, SeqMan Ultra provides a fast method of moving from one variant to the next along the sequences. Start by clicking the magnifying glass tool in the upper right of the Alignment view:
From the Search menu, choose Variant; then use the green arrows to search for variants upstream and downstream of the current cursor position.
Viewing and working with variants in a tabular display:
- SeqMan Pro – Open the Variants table by selecting a contig in the Project window and choosing Variant > Variant Report. Table columns can be added or removed by right-clicking on the table and choosing Show/Hide Column. The types of features included in the table are controlled using the Filter button. SNPs can be marked as putative/confirmed/rejected by clicking in the SNP column.
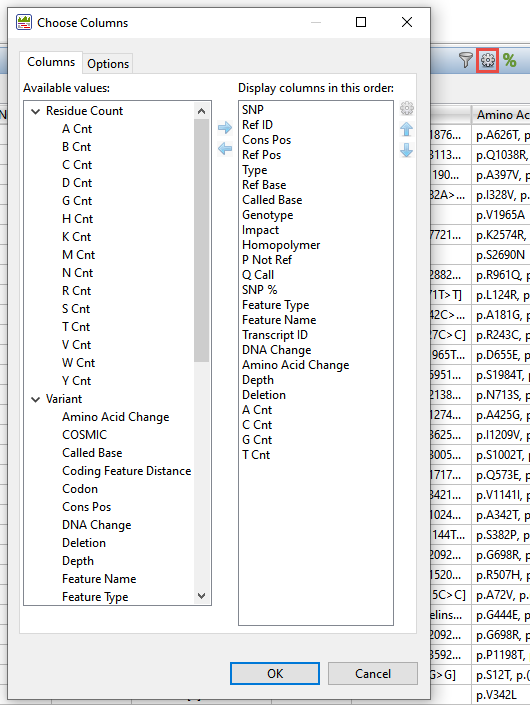
- SeqMan Ultra – Open the Variants view by selecting a contig in the Explorer panel and choosing View > Variants or by using the tool to the right of the Explorer panel.
As in SeqMan Pro, SNPs can be marked as putative/confirmed/rejected by clicking in the SNP column.
From within the Variants view, table columns can be added or removed using the “gear” tool in the top right of the view.
To filter variants shown in the view, use the “filter” tool in the top right of the view to open a filter dialog.
Need more help with this?
Contact DNASTAR