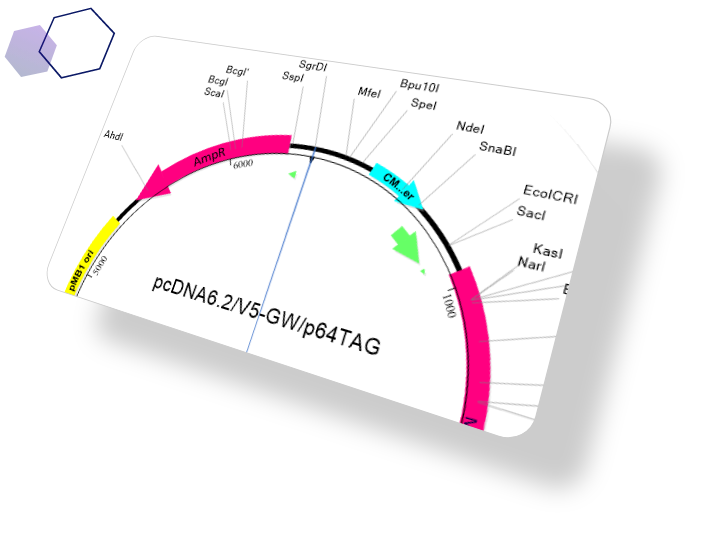
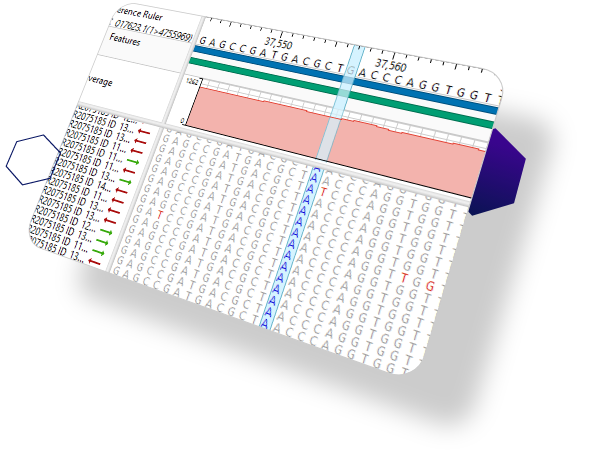
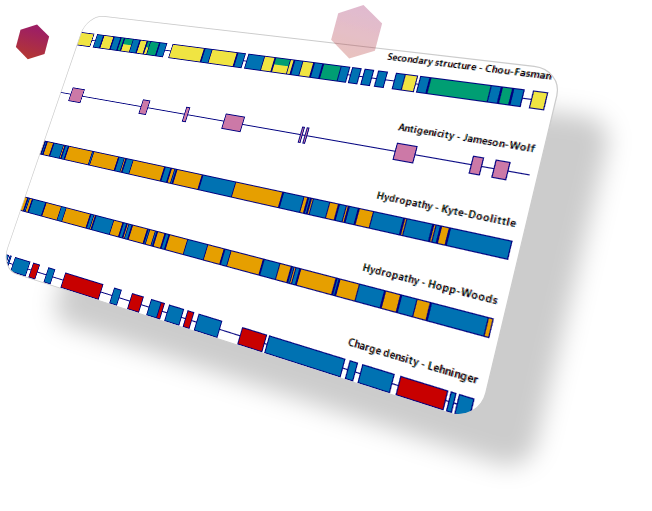
The most comprehensive sequence analysis tools on the market
DNASTAR software is trusted by tens of thousands of researchers around the world because of its well-deserved reputation for accuracy, versatility, and ease of use.
Our cutting-edge applications expertly handle dozens of popular bioinformatics workflows, including molecular cloning and primer design; Sanger, NGS, and long-read data assembly and analysis; multiple sequence alignment; and transcriptome analysis.
Software that covers all your sequence analysis needs
Available as an all-inclusive solution or as any of three packages designed to align with specific research objectives.
Protein Modeling Solutions
Understanding the structure of a protein is critical to understanding protein function. Our Nova Applications integrate with Lasergene and incorporate the biophysical properties of the protein sequence into comprehensive structure prediction and analysis.
Application Guides
Identification and Characterization of Viral Strains
This guide leads you through the process of viral genome sequencing and characterization: from choosing a sequencing strategy for templated or de novo assembly, through downstream analysis. It also shows the results from our survey, including a discussion of the top challenges faced by virologists performing this important work.

7 Steps for Human Variant Analysis
In this guide, we discuss some of the challenges involved in human variant analysis and explore some of the solutions available for addressing these challenges. You will learn about important considerations to keep in mind throughout the process, whether you are working with a core facility, a bioinformatics group, or doing variant analysis on your own.
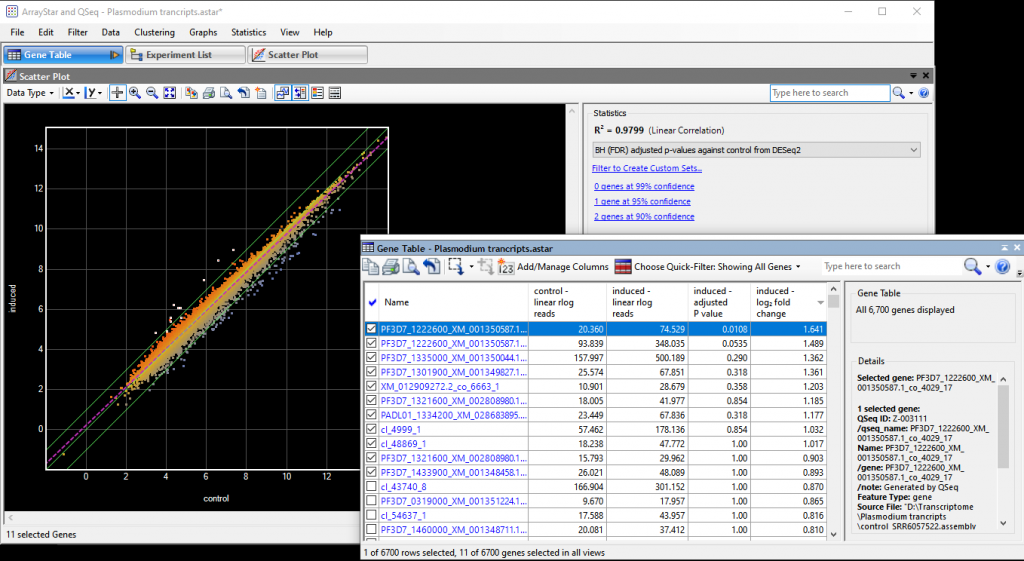
Successful De Novo Transcriptome Sequence Assembly Using RNA-Seq Data
De novo transcriptome analysis involves assembling cDNA sequences together—without the aid of a reference sequence—and then analyzing the resulting transcriptome. This technique is frequently used to study non-model organisms but is also valuable for organisms whose genome is known. Whether you are experienced with transcriptome analysis or thinking about trying it for the first time, this guide highlights factors to keep in mind when using this approach.
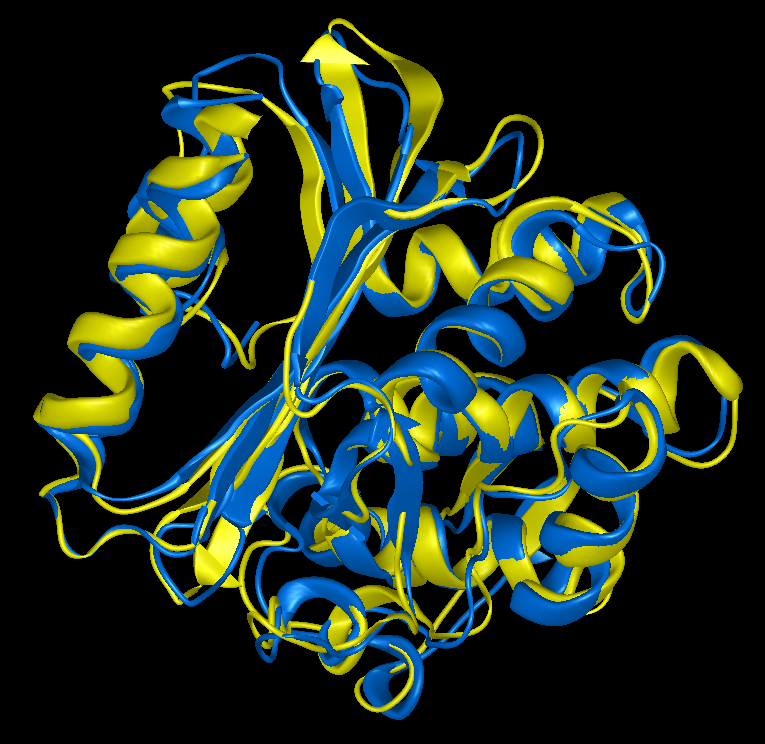
Protein Structure Prediction with NovaFold AI and NovaFold AI-Multimer
Of hundreds of single-chain protein structure prediction algorithms that have been objectively tested, the undisputed champion is AlphaFold 2, an artificial-intelligence based algorithm developed by Google's DeepMind. An extension of the algorithm, called AlphaFold-Multimer, was released a year later and is optimized for predicting the structures of protein-protein complexes. This guide describes the steps involved in determining protein structures, the advantages and challenges of using open source AlphaFold 2 and AlphaFold-Multimer, and describes how NovaFold AI and NovaFold AI-Multimer provide an easier way to use these powerful algorithms.
- Viral Genome Analysis
-

Identification and Characterization of Viral Strains
This guide leads you through the process of viral genome sequencing and characterization: from choosing a sequencing strategy for templated or de novo assembly, through downstream analysis. It also shows the results from our survey, including a discussion of the top challenges faced by virologists performing this important work.
- Human Variant Detection
-

7 Steps for Human Variant Analysis
In this guide, we discuss some of the challenges involved in human variant analysis and explore some of the solutions available for addressing these challenges. You will learn about important considerations to keep in mind throughout the process, whether you are working with a core facility, a bioinformatics group, or doing variant analysis on your own.
- Transcriptome Analysis
-

Successful De Novo Transcriptome Sequence Assembly Using RNA-Seq Data
De novo transcriptome analysis involves assembling cDNA sequences together—without the aid of a reference sequence—and then analyzing the resulting transcriptome. This technique is frequently used to study non-model organisms but is also valuable for organisms whose genome is known. Whether you are experienced with transcriptome analysis or thinking about trying it for the first time, this guide highlights factors to keep in mind when using this approach.
- Protein Structure Prediction
-

Protein Structure Prediction with NovaFold AI and NovaFold AI-Multimer
Of hundreds of single-chain protein structure prediction algorithms that have been objectively tested, the undisputed champion is AlphaFold 2, an artificial-intelligence based algorithm developed by Google's DeepMind. An extension of the algorithm, called AlphaFold-Multimer, was released a year later and is optimized for predicting the structures of protein-protein complexes. This guide describes the steps involved in determining protein structures, the advantages and challenges of using open source AlphaFold 2 and AlphaFold-Multimer, and describes how NovaFold AI and NovaFold AI-Multimer provide an easier way to use these powerful algorithms.
Who Uses DNASTAR Software?
Whatever your field of research—from archaeology to zoopathology—DNASTAR bioinformatics software is the ultimate solution to simplify your life. Just ask any of our thousands of loyal customers around the world!

Workflows to support
your research
We offer a wide variety of workflows for comprehensive sequence analysis; assembly and analysis of NGS, long-read, and transcriptomics data; and protein sequence analysis and structure prediction.
We know you’re busy.
We can help.
While we aim to make our software easy to use, some researchers don’t have a moment free to learn bioinformatics software. That’s why we offer genomic services for assembling, analyzing, and annotating genomic sequencing data, as well as in silico protein analysis services for antibody design, protein structure prediction, and custom pipeline integration.

Get the Latest
Find out about upcoming webinars, news, and product updates.