NovaDock
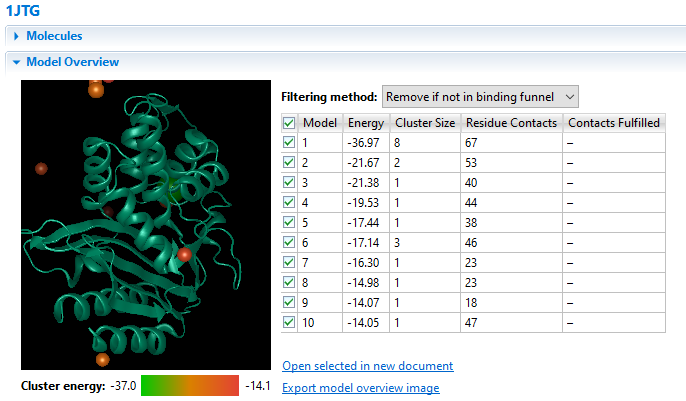
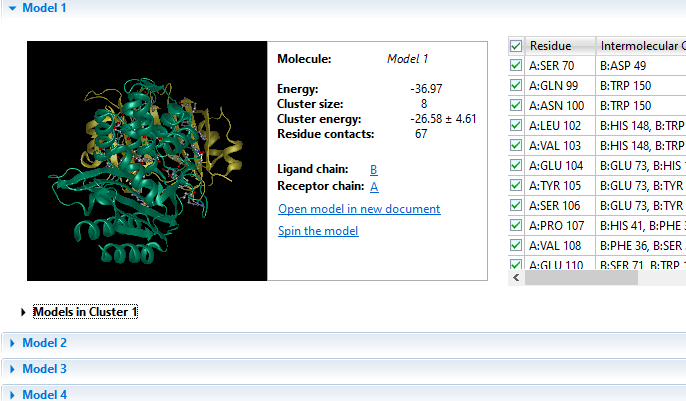
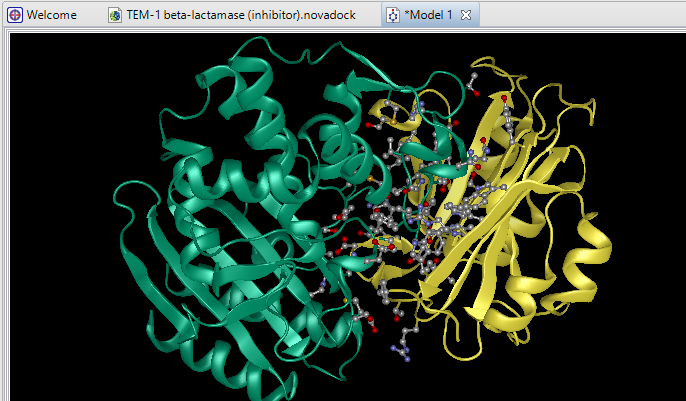
Due to its complex nature, accurately modeling a protein-protein docking interaction can often prove to be very challenging. NovaDock molecular docking software offers the ability to predict protein-protein docking interactions for any two binding partners utilizing SwarmDock, one of the top algorithms validated in the CAPRI blind docking experiment. Utilizing the SwarmDock algorithm, NovaDock explores protein flexibility when docking, resulting in more accurate predictions. Simply provide your ligand and receptor PDB or structure files, proposing specific residue contacts between the binding partners if desired. NovaDock will then predict the three-dimensional structure of the macromolecular complex and provide energy score, cluster size, and number of ligand contacts for each model for analysis in Protean 3D.
Resources
Please see our resources below for more information on NovaDock molecular docking software.
Protein-Protein Docking Workflow
How Can DNASTAR’s Protein Tools Help You?
High-Resolution in silico Protein Structure Prediction and Docking
Why Structure Prediction Matters
NovaDock Help
FAQs
Do I need a Lasergene license to run NovaDock molecular docking software?
Yes. A Lasergene Protein license is required to run NovaDock. Protean 3D, included in Lasergene Protein, provides the interface for running NovaDock predictions and analyzing the results.
Do you offer a free trial of NovaDock?
One prediction for each of our Nova Applications is included in our Lasergene free trial. We also offer free, no-obligation, personalized demos of any of our Nova Applications with one of our scientists on staff, tailored to your specific research objectives.
What prediction method does NovaDock use?
NovaDock’s algorithm is based on SwarmDock, developed in Dr. Paul Bates’ laboratory at the Cancer Research UK’s London Research Institute, and ongoing at the Francis Crick Institute. NovaDock does not use a library of templates, but instead makes docking predictions based on a type of energy calculation known as “particle swarm optimization.”