If you are viewing an assembly created in SeqMan NGen, you can add a track to the Analysis view showing each of the aligned reads in the selected experiment using the method described in Work with Tracks. Briefly, you can display an Alignment track by opening the Tracks panel and checking one or more of the boxes to the left of Analysis body > Alignment > (sample names).
Depending on the zoom level in the view, the Alignment track will show base letters (above) or green and red arrows representing forward and reverse reads, respectively (below).
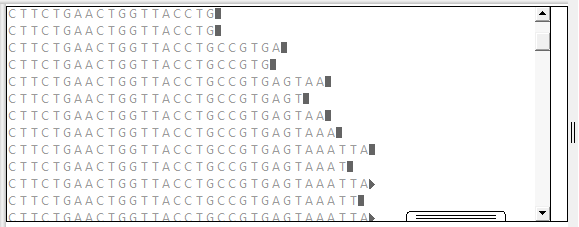
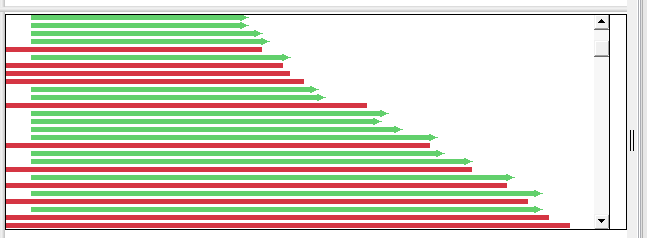
To resize the track vertically, use the slider that appears if you hover over the bottom middle of the track data. You can also resize the track and change the amount of white space above/below the track using sliders in the Layout section.
You can also display quality score information on the Alignment tracks as shown in the image below.
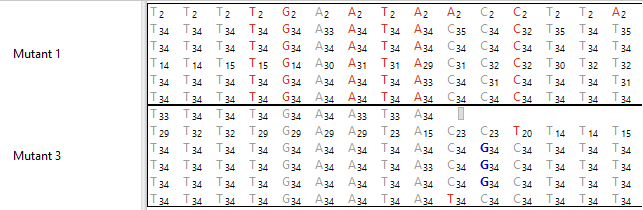
Unlike most other track types, Alignment tracks have their own tools. To see the Alignment track tools, first apply an Alignment track to the Analysis view, then select the Alignment track in that view. The toolbar will pop up at the top-left of the track. You can use your mouse to grab the popup on its left end and drag it to any desired location in the Analysis view.
| Tool name | Image | Purpose |
|---|---|---|
| Quality scores |  |
Choose between Don’t show scores and Show scores. Quality scores are only available for assemblies created using Sanger (.abi) reads. |
| Export data |  |
See Export data to a file for detailed instructions. |
| (Hamburger menu) |  |
Choose Tracks to open and focus on the Tracks panel. Choose Layout to open the Layout section for the Alignment track. |
| Close popup |  |
Closes the tool popup. |
To change Alignment track options:
Select the Alignment track in the Tracks panel and then click the Options tab to expand the Options section.
- Use the lowermost Read Layout menu to choose One sequence per line or One or more per line. If you choose One sequence per line, only one sequence will appear in each line, preceded or followed by blank space before and after the sequence. If you choose One or more per line, GenVision Pro saves vertical space by putting any sequences that do not interfere/overlap with another sequence on the same row as another sequence. These sequences are placed in areas that would otherwise have consisted only of blank space.
- Use the lowermost Quality Score menu to choose Don’t show scores, Show scores, or Average scores.
- Click
if you wish to return to the default values.
Need more help with this?
Contact DNASTAR



 if you wish to return to the default values.
if you wish to return to the default values.