The Analysis view shows tracks for a single sequence. The view is empty unless the session contains only one sequence; or until a single sequence has been chosen in the Overview or Explorer panel, or by using the navigation tools or Places panel bookmarks. At that point, the Analysis view appears automatically and is populated with data.
To access the Analysis view at any time, click on the Analysis tab, the dark blue tab in the image below, or use View > Analysis.
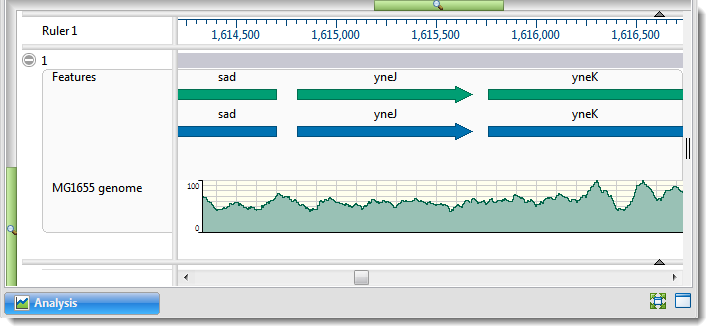
By default, the Analysis view displays a header and footer, along with a sequence and its corresponding data tracks. Tracks in the Analysis view can have different appearances depending on the zoom level.
The following table shows tasks associated with the Analysis view:
| Task | How To |
|---|---|
| Display the search tools | To display the search tools in the upper left of the Analysis view, use the Find tool (  ). ). |
| Sort constituent sequences | To sort read files (the files in the left margin of the view) according to the selected criteria, use the Sort reads tool ( ). ).
|
| Adjust the zoom level | Use the Compact text tool ( ) to adjust the zoom level to maximize the number of bases while keeping the text legible. Use the Restore default zoom tool ( ) to adjust the zoom level to maximize the number of bases while keeping the text legible. Use the Restore default zoom tool ( ) to adjust the zoom level to the (wider) default setting, making the text more comfortable to read. ) to adjust the zoom level to the (wider) default setting, making the text more comfortable to read. |
| Export data | Use the Export data tool ( ) and choose between Export Aligned Sequences and Export Consensus. This tool acts as a shortcut to the File > Export Data > Aligned Sequences and File > Export Data > Consensus commands and opens the same popup dialogs. See Export data to a file for information on using the dialog. ) and choose between Export Aligned Sequences and Export Consensus. This tool acts as a shortcut to the File > Export Data > Aligned Sequences and File > Export Data > Consensus commands and opens the same popup dialogs. See Export data to a file for information on using the dialog. |
| Export image | Use the Export image tool ( ). This tool acts as a shortcut to the File > Export Image > Analysis command. See Export an image of a view for more information. ). This tool acts as a shortcut to the File > Export Image > Analysis command. See Export an image of a view for more information. |
| Change the color scheme, font, or layout | Use the Style panel’s Sequence section and/or Analysis section. |
| Show/hide the header & footer | Click on the small dark gray arrows near the upper right corners of each section.  |
| Show a vertical guideline | Click Analysis > Show Guideline. The presence of a check mark directs GenVision Pro to display the guideline. A vertical guideline is displayed by default, and is a navigational aid to help in guiding your eye between sequences and tracks.  |
| Hide a vertical guideline | Click Analysis > Show Guideline. The absence of a checkmark means that only an abbreviated version of the guideline will remain visible in the header and footer. |
Need more help with this?
Contact DNASTAR


