The Per-Template Report is one of the reports available in the Reports views and is available for some types of .assembly projects. This report displays the contents of a file written by SeqMan NGen’s XNG assembler called perTemplateResults.txt. The report’s table can be customized to display a wide variety of data columns in any desired order.
To display this report, select a row in the Experiments section of the Explorer panel and click the Show Reports () tool or use the View > Reports command. Then use the drop-down menu near the top left of the table to choose per-Template.

To export the table, click on the Export tool ( ).
To select which columns to display in this table, or to rename or reorder the columns, click the Choose or rearrange columns (
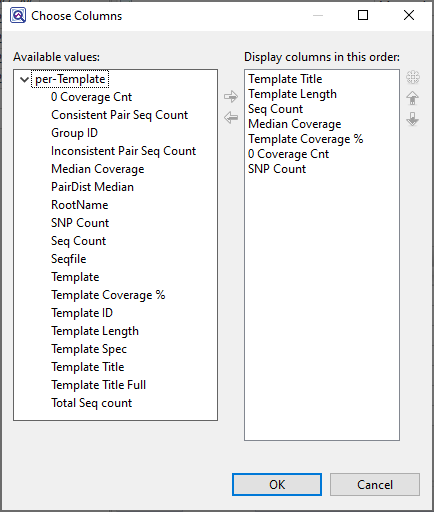
Available columns are on the left, while currently-applied columns are on the right.
- To add a column to the display, select its name on the left and press the right arrow key to move it to the right.
- To remove a column from the display, select its name on the right and press the left arrow key to move it to the left.
- To change the order of displayed columns, select the column name you wish to move on the right, then use the up/down arrows to place it in the desired order.
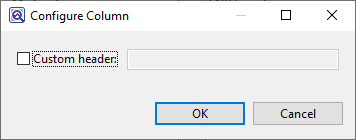
- To create a custom header for a displayed column, select its name on the right, then choose the Configure column tool (
). In the popup dialog, type in the desired name and press OK.
Descriptions of each column appear below in alphabetical order.
| Column Name | Description |
|---|---|
| 0 Coverage Cnt | The number of bases in the reference sequence with zero coverage in the alignment. |
| Consistent Pair Seq Count | For paired end sequencing (i.e. Illumina), the number of aligned read pairs that are correctly oriented and within the specified minimum and maximum distances. |
| Group ID | Experiment Name assigned by the user when setting up the assembly in SeqMan NGen. |
| Inconsistent Pair Seq Count | For paired end sequencing (i.e. Illumina), the number of aligned read pairs that are incorrectly oriented and/or outside the specified minimum and/or maximum distances. |
| Median Coverage | The median depth of coverage across the individual contig. |
| PairDist Median | The median distance between paired reads in the data set. |
| RootName | The file name and complete file path of the template file without the .bam extension. |
| SNP Count | The number of potential SNPs that were identified. |
| Seq Count | The number of sequences in the template file. |
| Seqfile | The file name and complete file path of the template file, including the .bam extension. |
| Template | The file name and complete file path of the template file-of-files, including the .template.fof extension. |
| Template Coverage % | The percentage of bases in the template covered by aligned sequence reads. |
| Template ID | Internal identifier of the template used by DNASTAR for trouble-shooting. |
| Template Length | The length of the template in base pairs. |
| Template Spec | File pathway to the reference sequence. |
| Template Title | The complete field of the reference sequence. |
| Template Title Full | (DNASTAR internal use only) |
| Total Seq Count | The total number of sequences in the layout for that template. Includes assembled and unassembled reads. Both halves of a split read are counted whether they aligned or not. |
Need more help with this?
Contact DNASTAR


