To translate one or more nucleic acid sequences into protein sequences using a specified translation table:
- Choose Tools > Batch Translate. The Project window opens with the Project and Options tabs active.
- In the Project tab, the sequences you wish to translate should be placed in the Sequences folder. To add sequences to a folder, select the folder, then right-click it and choose Import. Select the desired sequence files, and press Open. Next, select a single sequence, or use Shift+click or Ctrl/Cmd+click to select multiple sequences.
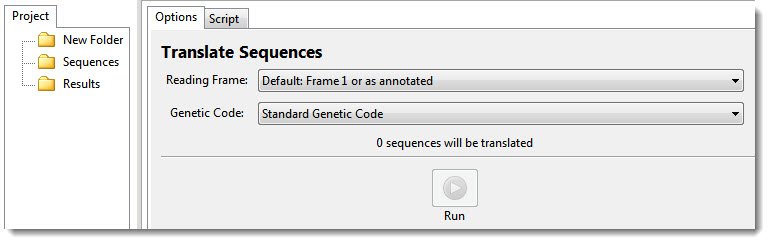
In the Options tab, the number of sequences selected on the left appears in the message: “‘n’ sequences will be translated.’
- In the Options tab, use the Reading Frame drop-down menu to choose the reading frame for the translation.
- Default: 1 or as annotated – Honor any existing /codon_start annotations in the input file(s); or otherwise, to begin at the first base.
- Ignore ‘/codon_start’ annotations – Begin translating at the first base and to ignore any /codon_start sequence annotations. Note that this selection is equivalent to the Default: 1 or as annotated selection (above) for any input sequence other than a .gbk or .seq file with /codon_start qualifiers.
- 1, 2 and 3 – Begin the translation at the first, second or third base, respectively, ignoring any /codon_start annotations. For an input sequence containing /codon_start qualifiers, selecting the start codon implicitly is similar to choosing Ignore ‘/codon_start’ annotations, but allows a specific frame to be set.
- Default: 1 or as annotated – Honor any existing /codon_start annotations in the input file(s); or otherwise, to begin at the first base.
- In the Genetic Code drop-down menu, choose a code for the translation. All NCBI and Lasergene tables are available, and are denoted by number and/or description. By default, the translation uses the genetic code identified in the source data (e.g., the GenBank /transl_table qualifier), if available; otherwise, it uses the standard genetic code (NCBI:1).
- Press the Run button.
If an input sequence was annotated, each CDS feature is translated separately, and any translation table and/or codon_start annotations are honored. If the input file was not annotated, or did not contain CDS features, the entire sequence is translated. Note that translation halts when there are fewer than 3 bases left. An “extra” base or two at the end will not be reflected in the output.
The new sequence(s) appear in the Project tab’s Results folder.
- (optional) If you wish to see the script that was used to run this process, press the Script tab.
- (optional) If you want to see the history of the run, including the location of the output file(s), press the History tab.
Need more help with this?
Contact DNASTAR


