If—when setting up the SeqMan NGen assembly—you chose Multi-sample or Multi-sample with replicates in the Input sequences screen, SeqMan Ultra displays a special version of the Alignment view. This version shows a “pseudo-consensus” where you can compare different strains, varieties, etc., and easily see which samples have variants.
To view multi-sample assemblies in the Alignment view:
- Open a multi-sample assembly.
- In the Explorer panel, select the contig of interest.
- Press the Show table of variants in selected contigs tool (
) to launch the Variants view.
Note that the Sample column contains the Experiment or Replicate name you specified on SeqMan NGen’s Input sequences screen.

- Double-click on a variant of interest to launch the Alignment view at that position. The view shows SNPs in blue text on an orange background, allowing you to easily see which samples have variants.
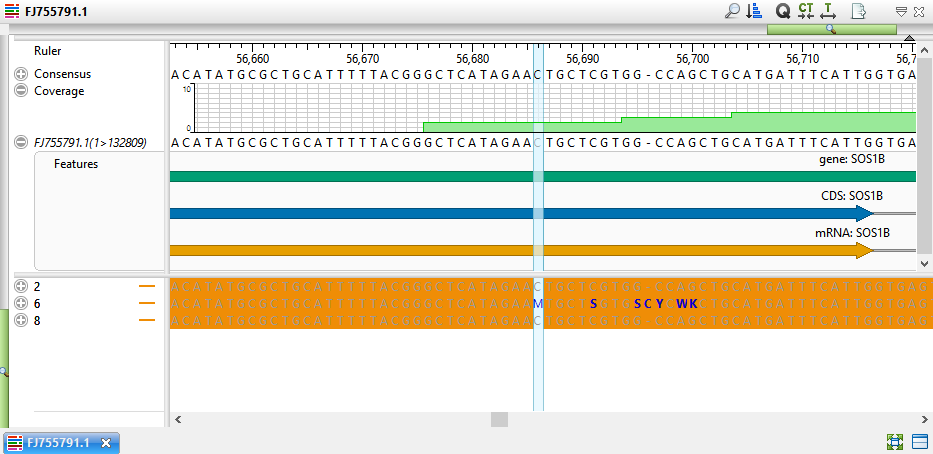
- If desired, click the plus signs to expand the samples and view the individual read sequences. You can also expand the read sequences in the same way to see trace data, if trace files were used in the assembly.

Need more help with this?
Contact DNASTAR



 ) to launch the
) to launch the