The Primer Design view is one of the ten is one of the ten views available in the Document window, and shares a sub-set of button tools with the other views. It also has its own set of tools.
To make it the active view (or one of two active views), click on the Primer Design tab ( 
The Primer Design view lets you design and analyze primers for a nucleotide sequence. Data in the Primer Design view appears in a header (featuring tools and details about the primers) and three distinct panes below that header. See Work with Primers for detailed information about performing tasks within this view.
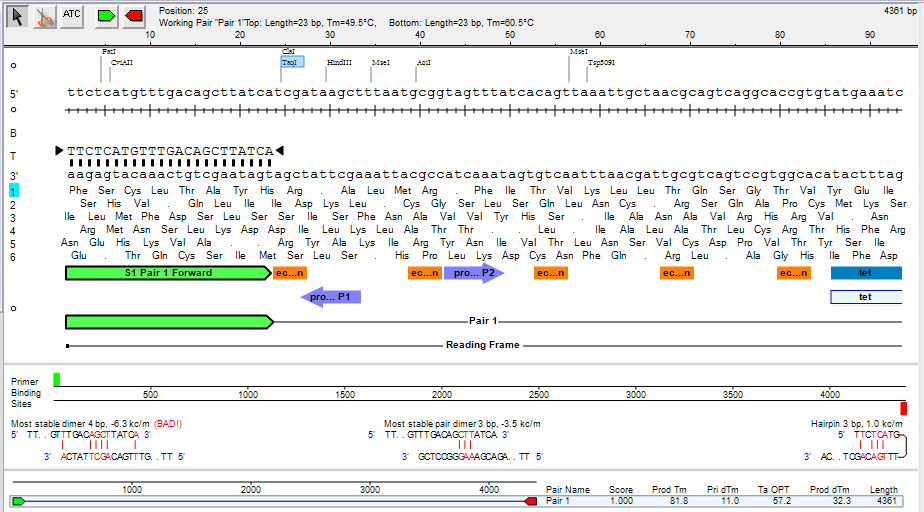
Sequence details in the header:
Details about the primers, if any, are displayed in the header of the Primer Design view, just to the right of the tools.
| Selection/Position | The current position of the cursor. If a range is selected, the length of the range and strand (forward or reverse) is specified, along with the sequence coordinates. |
| Working Pair | The name of the currently selected primer pair. |
| Forward | The length and melting temperature of the forward primer. This value is updated automatically if settings are changed. |
| Reverse | The length and melting temperature of the reverse primer. This value is updated automatically if settings are changed. |
| Sequence Length | The total length of the sequence. This value is shown in the top right corner of the header. |
Residue pane:
Displays the base level nucleotide sequence, restriction sites, translations, and features, as well as the currently selected primer sequence. This pane can be used to modify your primers.
Once you have done this, a green arrow represents the forward strand primer; red indicates the reverse strand primer. Primer sequences are displayed in 5’ to 3’ orientation, and include modifications that you have made. Edited nucleotides appear in lowercase.
Mispriming pane:
Shows primer binding sites, and the most stable conformation of dimers, pair dimers, and hairpins, if they exist. As the current primer pair is changed, the information displayed in the Mispriming Pane is updated automatically.
At the top of this pane, Primer Binding Sites displays where the primer binds to the template as well as any additional sites the primer may bind to on either strand. Any sites on the template matching at least the 12-mer at the 3’ end of the primer are displayed. Primers on the top strand are indicated in green; primers on the reverse strand are shown in red.
Beneath Primer Binding Sites, the most stable conformation of dimers, pair dimers, and hairpins are shown, if they exist. To calculate this, SeqBuilder Pro locates secondary structure interactions using the nearest-neighbor model for all three types of duplexing. For self and pair dimer duplexing, oligonucleotide sequences are compared at every potential overlapping position by aligning sequences and advancing one residue at a time.
- Most stable dimer - Displays the primer binding to itself in the most stable conformation, if there is one.
- Most stable pair dimer - Displays the primer binding to the opposite strand primer in the most stable conformation, if there is one.
- Most stable hairpin - Displays the primer binding to itself by folding in the most stable conformation, if there is one.
Alternate pairs pane:
Displays the list of primer pairs discovered by your search, along with additional information for each pair, including product length, melting temperatures, and a quality score. The following information is displayed in columns for each pair:
| Name/Abbrev. | Description |
|---|---|
| Pair Name | The name of the primer pair. |
| Score | The quality score assigned to the primer pair. |
| Prod Tm | The melting temperature of the product. |
| Pri DTm | The difference in melting temperature between the primers in the pair. |
| Ta OPT | The optimal annealing temperature for the primer pair. |
| Prod dTm | The difference in melting temperature between the product and the lower primer Tm*.* |
| Length | The length of the product. |
Need more help with this?
Contact DNASTAR


