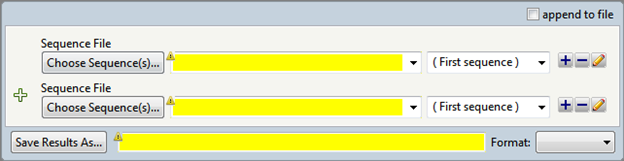
The Concatenate Sequences template, located in the Templates panel, initially prompts you to enter two sequences and to choose the desired file output and location. You can also add additional items to be concatenated (e.g., additional sequences, sequence text strings) using the right-click menus or any of the Sequences section tools that start with the word “Add.
Later, when you run the script, SeqNinja will concatenate all combinations of the sequence(s) and/or text string(s) you entered and output the results to the selected file. The following rules apply:
- Any ranges added to this step are re-evaluated for each member of the set to which they are applied.
- The plus (+) operator concatenates, or links together, corresponding pairs in the left and right sequence sets.
- When one of the rows is a singleton (e.g. a literal such as “ATG”), the concatenation operation results in prepending or appending that single sequence to every sequence in the other row(s).
- In the Choose Sequence(s) button row, choose the sequences you wish to join (see Add and modify a sequence).
- Add additional sequences or modifiers, if desired, using the right-click menu or any of the Sequences section tools that start with the word “Add.”
- In the Save Results As area, choose the name and location in which to save the output (see Specify output format and location).
Examples:
Simple concatenation example:
Input sequence 1: GGAGTCTCCC
Input sequence 2: ATTGATTACA
Output sequence: GGAGTCTCCCATTGATTACA
To add “ATG” to the beginning and “TAG” to the end of every sequence in a file:
Input sequence 1: ATG
Input sequence 2: mysequences.fasta
Input sequence 3: TAG
Output sequences: ATGcggtaaatcTAG, ATGgttaccaaTAG, etc.
To concatenate sub-ranges of sequences in a file:
Input sequences: Sub-ranges 1-3 and 4-6 of the file two.fasta, which contains the following two sequences: gttacgtcgacgcgggcgtg and GTTACGTCGACGCGGGCGTG.
Output sequences: gttacg, GTTACG
Need more help with this?
Contact DNASTAR