The Split Sequence Set tool, located in the Other section of the Toolkit panel, separates multi-sequence files into multiple files, each consisting of a single sequence.
Note: Sequences that have been split may later be combined in a common file using the Collect Sequences tool.
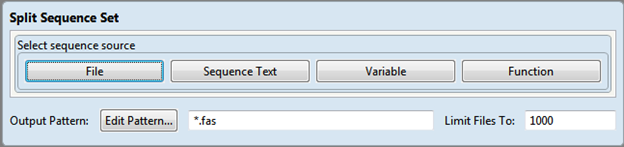
- This dialog requires you to select a sequence source. See Write to Results File for information on using the File, Sequence Text, Variable and Function buttons.
- To specify the required filename pattern, click the Edit Pattern button. See Use File Patterns for detailed information.
- If desired, enter a value in the Limit Files To box to limit the number of sequence files that can be generated during a run. The default is 1000.
If you have specified “full” file conversion, each sequence file that is generated may be accompanied by auxiliary files, as well.
Example:

The input file, 454Reads.fna, consists of 13 sequences in a multi-sequence file format.
After running the script, the first ten sequences in 454Reads.fna are saved as individual sequence files in .fasta format.
Need more help with this?
Contact DNASTAR


