Lasergene Genomics
Lasergene Genomics stands apart in the fields of genomics and transcriptomics. Powered by SeqMan NGen, our revolutionary and easy-to-use assembler, the software enables you to set up complex genomic sequencing projects in mere minutes, and automates tasks that typically require extensive manual intervention in other software packages.
Intuitive project setup and analysis, combined with our patented assembly algorithms, allow you to assemble and align your NGS data with unsurpassed ease and speed, so that you can focus on the results. No more switching between software tools to assemble sequences, identify important variants, and determine differentially expressed genes. Everything you need is here.
Lasergene Genomics Workflows
Lasergene Genomics Applications
Genomic Sequence Assembly
SeqMan NGen enables you to set up complex genomic sequencing projects in mere minutes, then assembles NGS and long read sequencing data with unsurpassed ease and speed.
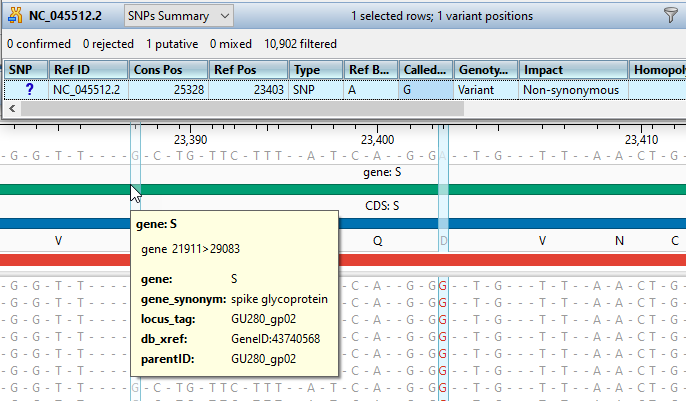
Gene Expression and Variant Analysis
ArrayStar’s advanced filtering tools and rich visualizations enable you to easily evaluate gene expression, and identify significant variants across multiple samples.
Sequence Assembly Analysis and Editing
SeqMan Ultra provides post-assembly analysis tools, such as evaluating coverage, analyzing variants within the context of the alignment, and contig editing.
Genomic Visualization Software
Visualize genomic sequencing results such as coverage and peak plots using highly-customizable images that are easily exported for collaboration or publication.
Cloud-Based Storage and Assemblies
DNASTAR Cloud Data Drive provides NGS data management. DNASTAR Cloud Assemblies* utilizes our cloud computing resources to align and assemble your NGS data, freeing up your local computer to do other things.
*Each Lasergene Genomics license includes five free DNASTAR Cloud Assemblies. Further cloud assemblies require an additional purchase.
Resources
Please see our resources below for more information on Lasergene Genomics.
Lasergene Genomics Overview
7 Steps for Human Variant Analysis
Choosing the best assembly strategy for your genomic sequencing data
RNA-Seq Assembly and Normalization Methods—an Interview with Dr. Carl-Erik Tornqvist
FAQs
What types of genome analysis can I do with Lasergene Genomics?
Lasergene Genomics includes all the applications you need for clinical research studies, de novo genome assembly, metagenomic assembly, advanced variant analysis, whole exome/genome sequencing, and transcriptomics studies such as ChIP-Seq, RNA-Seq, and de novo transcriptome assemblies.
Which applications are included with a Lasergene Genomics license?
Lasergene Genomics includes SeqMan NGen, SeqMan Ultra, ArrayStar, and GenVision Pro, as well as five DNASTAR Cloud Assemblies.
Do I need a powerful computer to run Lasergene Genomics?
Not necessarily. To run Lasergene Genomics locally, hardware requirements vary based on the type of projects you plan to do. Please see our technical requirements for details.
Because performing…
Not necessarily. To run Lasergene Genomics locally, hardware requirements vary based on the type of projects you plan to do. Please see our technical requirements for details.
Because performing large scale NGS alignments or de novo sequence assemblies can require significant hard disk space and memory, we offer DNASTAR Cloud Assemblies, so that you can easily utilize our cloud computing resources to align and assemble your NGS data, using any laptop or desktop computer that meets our minimum technical requirements.
Every free trial and purchase of Lasergene Genomics includes five free DNASTAR Cloud Assemblies.
How much does Lasergene Genomics cost?
We offer both academic and commercial pricing for convenient annual licenses of Lasergene Genomics, which include upgrades and unlimited access to our technical…
We offer both academic and commercial pricing for convenient annual licenses of Lasergene Genomics, which include upgrades and unlimited access to our technical support team.
We also offer floating licenses for networks, and site licenses that can be shared with an entire organization. For pricing on network or site licenses, please request a quote.
Can I try Lasergene Genomics before I buy it?
Yes! We offer free, no-obligation trials of our complete DNASTAR Lasergene package.
Because genomics projects can be complex and sometimes overwhelming, we also encourage…
Yes! We offer free, no-obligation trials of our complete DNASTAR Lasergene package.
Because genomics projects can be complex and sometimes overwhelming, we also encourage you to sign up for a free, one-on-one demonstration, customized to your specific research objectives and led by one of the scientists on staff.
Compare DNASTAR Lasergene Packages
| MOST POPULARDNASTAR Lasergene | ||||
| Lasergene Molecular Biology | Lasergene Genomics | Lasergene Protein | ||
|---|---|---|---|---|
| Included Applications | ||||
| SeqBuilder Pro | ||||
| SeqMan Ultra | ||||
| MegAlign Pro | ||||
| GeneQuest | ||||
| GenVision | ||||
| SeqNinja | ||||
| SeqMan NGen | ||||
| ArrayStar | ||||
| GenVision Pro | ||||
| Protean 3D (+1 prediction per Nova Application) |
||||
| DNASTAR Navigator | ||||
| Supported Workflows | ||||
| Integrates with | ||||
| See Pricing | See Pricing | See Pricing | See Pricing | |