Lasergene Molecular Biology
Every molecular biologist needs reliable sequence analysis software for everyday tasks such as editing and aligning sequences and designing workable primers and clones. And while these tasks may be routine, their success impacts everything else downstream. That’s why it’s so important to have dependable, easy-to-use tools that get it right, every time.
Lasergene Molecular Biology has you covered for important sequence analysis tasks including multiple sequence alignments, PCR primer design, in silico cloning, and Sanger sequence assembly. Now you can reach your analysis goals more quickly with insightful features like batch editing, automated sequence annotation, access to a meticulously annotated plasmid vector map catalog, and full integration with NCBI databases. Lasergene Molecular Biology goes beyond the basics, delivering truly remarkable sequence analysis software that you can rely on.
Lasergene Molecular Biology Workflows
Lasergene Molecular Biology Applications
Sequence Editing, Cloning, and Primer Design
SeqBuilder Pro provides sequence analysis tools for effortless sequence editing and annotation, plasmid map creation, in silico cloning and PCR primer design.
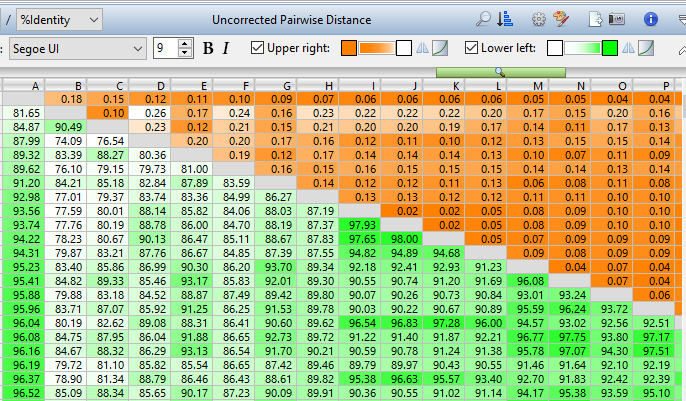
Multiple Sequence Alignments
MegAlign Pro performs multiple sequence alignments quickly and easily, then guides you through the post-alignment process, including thorough phylogenetic analysis.
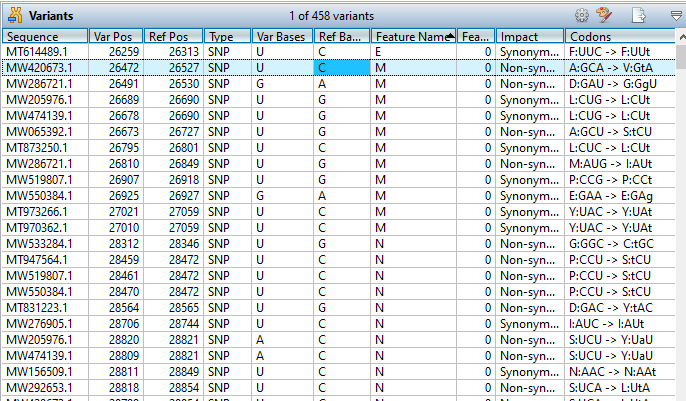
Sanger Sequence Assembly and Analysis
SeqMan Ultra accurately assembles Sanger ABI sequencing data with or without a reference, and includes an easy-to-use interface for previewing and trimming trace data.
Gene Discovery and Gene Annotation
GeneQuest offers a comprehensive set of analytical methods to facilitate finding and annotating the genes and regulatory elements on a sequence.
Automated Sequence Editing Utility
SeqNinja makes it easy to edit large groups of sequences and annotations simultaneously using customizable templates for functions such as translation and format conversion.
Genome Illustration Utility
GenVision supports the generation of customizable publication-quality graphics for linear or circular genome mapping.
FAQs
What types of sequence analysis can I do with Lasergene Molecular
Biology?
Lasergene Molecular Biology includes all the applications you need for plasmid map creation, primer design, in silico cloning, multiple and pairwise sequence alignments, Mauve genome alignments,…
Lasergene Molecular Biology includes all the applications you need for plasmid map creation, primer design, in silico cloning, multiple and pairwise sequence alignments, Mauve genome alignments, phylogenetic analysis, Sanger sequence assembly, gel electrophoresis simulation, and sequence editing and annotation.DNASTAR Lasergene is our complete software package which includes Lasergene Molecular Biology, Lasergene Protein, and Lasergene Genomics.
How much does Lasergene Molecular Biology cost?
We offer both academic and commercial pricing for convenient annual licenses of Lasergene Molecular Biology, which include upgrades…
We offer both academic and commercial pricing for convenient annual licenses of Lasergene Molecular Biology, which include upgrades and unlimited access to our technical support team. Special pricing is available for current students.
We also offer floating licenses for networks, and site licenses that can be shared with an entire organization. For pricing on network or site licenses, please request a quote.
Can I try Lasergene Molecular Biology before I buy it?
Yes! We offer free, no-obligation trials of our complete DNASTAR Lasergene package. During your trial period, please feel free to contact us with questions!
Compare DNASTAR Lasergene Packages
| MOST POPULARDNASTAR Lasergene | ||||
| Lasergene Molecular Biology | Lasergene Genomics | Lasergene Protein | ||
|---|---|---|---|---|
| Included Applications | ||||
| SeqBuilder Pro | ||||
| SeqMan Ultra | ||||
| MegAlign Pro | ||||
| GeneQuest | ||||
| GenVision | ||||
| SeqNinja | ||||
| SeqMan NGen | ||||
| ArrayStar | ||||
| GenVision Pro | ||||
| Protean 3D (+1 prediction per Nova Application) |
||||
| DNASTAR Navigator | ||||
| Supported Workflows | ||||
| Integrates with | ||||
| See Pricing | See Pricing | See Pricing | See Pricing | |
Resources
Please see our resources below for more information on Lasergene Molecular Biology.
Simplifying Multiple Sequence Alignment and Analysis with MegAlign Pro
Best Practices for Automated Virtual Cloning
Two ways to find the best MegAlign Pro multiple sequence alignment method for your data
Lasergene Molecular Biology Overview