Contig Information is one of three Contig Report views that may be available, depending on the assembly type that is currently open. The information in this view depends on the type of assembly (i.e., .sqd vs. .assembly ; templated vs. non-templated). Some parameter descriptions are shown in the table below:
For information on accessing or performing tasks in this view, see Contig Report View.
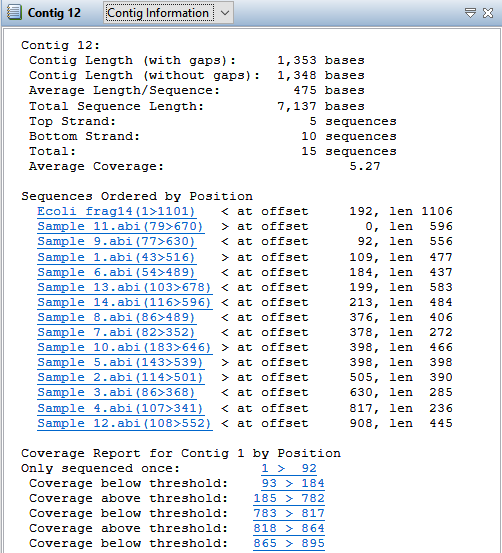
| Parameter | Description |
|---|---|
| Contig Length (with gaps) | The length of the consensus sequence. This length reflects gaps inserted into the individual sequences and any editing you have done to the contig. |
| Contig Length (without gaps) | The length of the consensus sequence without gaps. |
| Average Length/Sequence | The quotient of Total Sequence Length and the number of sequences in a contig. |
| Total Sequence Length | The sum of all individual sequence lengths. This length reflects gaps inserted into individual sequences. |
| Top Strand | The number of sequences assigned to the top strand of the consensus sequence. |
| Bottom Strand | The number of sequences assigned to the bottom strand of the consensus sequence. |
| Total | The total number of sequences used to construct a contig. |
| Average (or Median) Coverage | The average (or median) number of reads over the entire length of the consensus. |
| Sequences ordered by position | Summarizes the offset position and length for each sequence in a contig. |
| Coverage report for Contig ‘n’ by Position | Summarizes coverage in four categories: only sequenced once, sequenced only on one strand, sequenced on both strands but below threshold, and sequenced on both strands and equal to or above threshold. Each region is ordered by its position in the contig, from left to right. The threshold value is set in Strategy Viewing and Coverage parameters. |
| Coverage Summary (in bases) | Summarizes the number and frequency of bases in each coverage category. |
Double click on hyperlinked text to open the Alignment view for the contig in which that sequence appears, with the position or range already selected.
Need more help with this?
Contact DNASTAR


