From the Variants view, you can open a filter dialog to specify which variants are shown in the table. To open the Variant Filter Criteria dialog, use the Variants > Filter command, press the Filter tool (
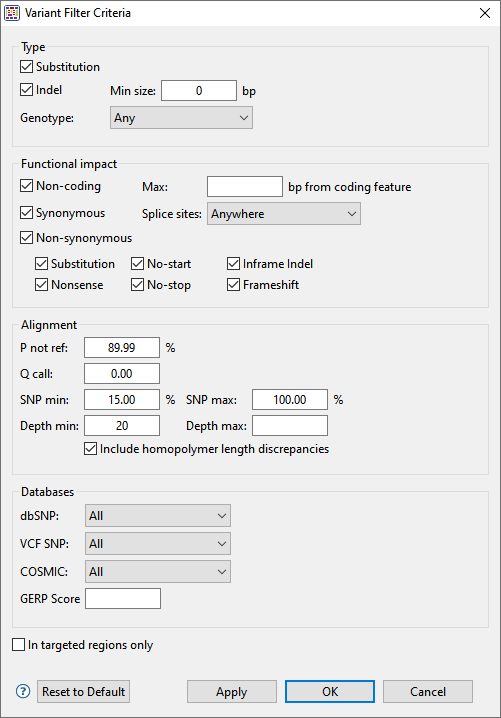
In the Type section:
Choose to display Substitutions and/or Indels by checking the boxes next to their name(s). If you want to see only variants greater than a certain length threshold, type that threshold into the Min size box.
Use the Genotype drop-down menu to limit the display to Annotated SNPs only or Novel SNPs only. To display both types, keep the default setting of Any.
In the Functional impact section:
Choose whether to display indels having the following functional impacts:
- Non-coding – The indel appears in a non-coding region of the sequence, and will therefore have no functional impact.
- Synonymous – The indel does not cause an amino acid change, and will therefore have no functional impact.
- Non Synonymous – Amino acid substitution only.
- Substitution – Any change from one nucleotide to another, regardless of effect or lack thereof.
- Nonsense – Amino acid to translational stop.
- No Start – A change that disrupts the start codon.
- No Stop – A change that converts a stop codon to an amino acid, and thereby extends the reading frame.
- Inframe Indel – An insertion or deletion within a coding region whose length is divisible by 3. The type is followed by the word Conservative if the indel occurs between two codons, and Disruptive if it occurs with a codon.
- Frameshift – An indel within a coding region and which is not a multiple of 3, thereby changing the reading frame.
- Substitution – Any change from one nucleotide to another, regardless of effect or lack thereof.
In the Alignment section:
- P not ref – The probability that the called base at this position is not the reference base. For coalesced variants, this value is equal to the minimum value of all “child” values. The minimum allowed value is 30%.
- Q call – The Phred-like quality score of the called genotype. It is a measure of the probability that the called genotype is correct.
- SNP min / SNP max – The minimum and maximum percentage of reads that should contain a SNP at a given position.
- Depth min / Depth max – The minimum and maximum depth of reads needed to include the SNP.
- Include homopolymer length discrepancies – If you are using Illumina or Sanger data, we recommend leaving this box checked. If using Ion Torrent data, we recommend unchecking the box. If you uncheck the box, SeqMan Ultra will remove all homopolymeric run length variants from the table.
In the Databases section:
- Use the dbSNP, VCF SNP and COSMIC drop-down menus to filter in/out subsets of variants from these databases.
- To filter out variants with a GERP score below a specified threshold number, enter that number in the text box.
- To vilter out variants from locations other than the targeted regions (i.e., from a .bed or manifest file used in the assembly), check the box next to In targeted regions only.
If you wish to return to default settings, click Reset to Default.
To save your settings, click Apply or OK, or simply press Enter.
Need more help with this?
Contact DNASTAR


