To access options affecting the display of pairwise alignments in the Pairwise view, click on the Style panel expand bar entitled Pairwise Alignment, or choose View > Style > Pairwise Alignment.
| Task | How To |
|---|---|
| Specify the wrapping style for rows | Use the Layout drop-down menu. Choose between:
|
| Visualize flanking segments of sequences that are not within the aligned segment | Check the Show context box. A local alignment reports the highest scoring contiguous segment of alignment between two sequences, even if the full extent of one or both of the sequences is not included in the final alignment. Local alignments are ideal for finding short sequences within a larger sequence. When you check Show context, the flanking segments appear in a dimmer color scheme than the alignment. As an example, consider a local alignment of a long sequence to a short one. Any positions that aren’t part of the alignment are considered to be context and are not displayed in the Pairwise view unless Show context is selected. In the case below, the segment “GGGC” in on the left of Seq1 is context, as is the trailing “TTAAAA”. The internal part of Seq2 that spans a gap in Seq1 might be considered unaligned, but is actual context, because it’s not internal to the alignment. Seq1 GGGC CCC -- TTA TTAAAASeq2 ---- CCC C C TTA ------ |
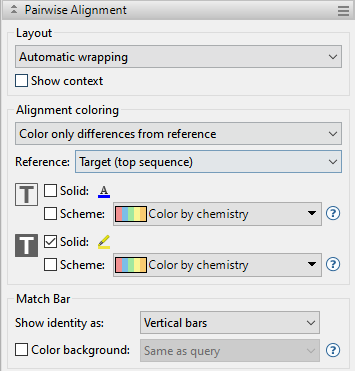
| Specify how to color the sequences and the consensus to facilitate visual comparisons | Use the Alignment coloring drop-down menu to s. Sequence comparisons can only be performed with aligned sequences. Choose between:
|
| Reference | Choose which sequence to use as the “Reference” in the pairwise comparison. Choices are Target (top sequence) or Query (bottom sequence). |
| Specify the color of sequence characters | Use the Foreground color tools to the right of  . To display the letters in a single solid color of your choice, check the upper box. Then click on the ‘A’ tool to the right of the checkbox and choose a color from the color picker. To instead choose a preset color scheme, check the lower box and make a selection from the drop-down menu. . To display the letters in a single solid color of your choice, check the upper box. Then click on the ‘A’ tool to the right of the checkbox and choose a color from the color picker. To instead choose a preset color scheme, check the lower box and make a selection from the drop-down menu.  Color scheme choices are described in Available color schemes. |
| Specify the background color (boxes around each letter) | Use the Background color tools to the right of  . To use a single solid color of your choice, check the upper box. Then click on the ‘highlighter pen’ tool to the right of the checkbox and choose a color from the color picker. To instead choose a preset color scheme, check the lower box and make a selection from the drop-down menu (see image in row above). Color scheme choices are described in Available color schemes. . To use a single solid color of your choice, check the upper box. Then click on the ‘highlighter pen’ tool to the right of the checkbox and choose a color from the color picker. To instead choose a preset color scheme, check the lower box and make a selection from the drop-down menu (see image in row above). Color scheme choices are described in Available color schemes. |
| Specify the background color for gaps | Use the Background color for gaps tools to the right of  . To use a single solid color of your choice, check the box next to Solid. Then click on the ‘highlighter pen’ tool to the right of the checkbox and choose a color from the color picker. If you do not wish to use a background color for gaps, check the Default box. . To use a single solid color of your choice, check the box next to Solid. Then click on the ‘highlighter pen’ tool to the right of the checkbox and choose a color from the color picker. If you do not wish to use a background color for gaps, check the Default box. |
| Specify the appearance of the match bar track | Use the Match bar section to customize display of the optional Match bar track. This track is used to show similarities and differences between the query and target sequences. These options are mirrored in the Match Bar track options section. Choices made in either location are instantly reflected in the other location.
|
| Match Bar Symbol | Interpretation | Weight table value |
|---|---|---|
| Vertical line | Match | 1 |
| Colon | Favorable mismatch | (0,1) |
| Period | Neutral mismatch | 0 |
| Space | Unfavorable mismatch | < 0 |
Need more help with this?
Contact DNASTAR



