This application supports local, global and semi-global pairwise alignment methods. Pairwise alignments can only be performed when two sequences, and only two sequences, have been selected. A common workflow is to first perform a multiple alignment on an entire group of sequences. From the resulting Tree view, two closely related sequences can then be further analyzed by selecting them and performing a pairwise alignment.
Pairwise alignment tutorials:
The following tutorials use free data that can be downloaded from the DNASTAR website:
- Try It! – Follow a multiple alignment with Global pairwise alignments
- Try It! – Align transcripts to genes using Local and Global pairwise alignments
- Try It! – Use a Local pairwise alignment to find a gene within a genome
To perform a pairwise sequence alignment:
- Decide which two sequences you want to align. The sequences can be any length (DNASTAR has successfully aligned protein sequences up to 35,000 bases in length), but both must belong to the same category: DNA/RNA or protein. If one sequence is significantly longer than the other, use drag & drop in the Sequences view or Overview to organize them such that the longer sequence is above the shorter sequence.
- Select the two sequences and choose Align > Pairwise or right-click on the selection and choose Align Pairwise. Dialog options vary depending on whether protein or nucleotide sequences were selected. An example is shown below.
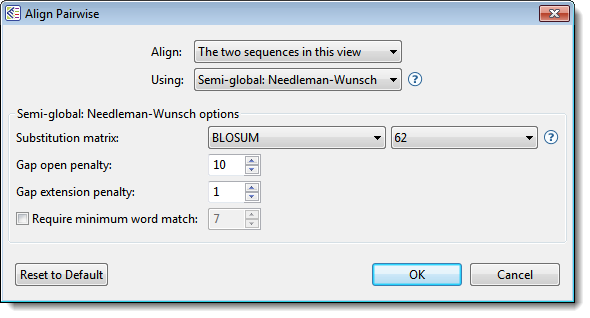
- Use the Align drop-down menu to specify which sequences to align.
- In the Using drop-down menu, choose the desired pairwise alignment method:
- Global alignment, represented in MegAlign Pro by the Global Needleman-Wunsch algorithm, includes the full length of both sequences, even if this requires padding one or more of the sequence ends with gaps. MegAlign Pro considers overhangs and underhangs created this way as unaligned context. One example of a situation where a global alignment is preferred over a local alignment is when there are multiple, but disjoint, segments of aligned sequence. Examples when a global alignment is a good choice: 1) aligning a CDS or mRNA sequence to a gene that contains introns; 2) aligning two sequences that differ because of the presence of large insertions, such as might be caused by transposable elements. In both cases, a local alignment is less likely to reflect the full alignment, especially if the lengths of the unalignable inclusions are long relative to the gap extension penalty.
- Semi-global alignment, represented by the Semi-global Needleman-Wunsch algorithm, is similar to global alignment, except the gaps placed at the ends of sequences are not penalized. A semi-global alignment might be more useful than global alignment in situations where long leading/trailing gaps might be suppressed in favor of a result that contains segments of aligned sequences punctuated by gaps.
- Local alignment, represented by the Local Smith-Waterman algorithm, reports the highest scoring contigous segment of alignment between two sequences, even if the full extent of one or both of the sequences is not included in the final alignment. Local alignments are ideal for finding a short sequence within a larger sequence. Flanking segments of sequences that are not within the aligned segment can be visualized in MegAlign Pro by checking the Show context box in the Pairwise Alignment section of the of Style panel.
- Global alignment, represented in MegAlign Pro by the Global Needleman-Wunsch algorithm, includes the full length of both sequences, even if this requires padding one or more of the sequence ends with gaps. MegAlign Pro considers overhangs and underhangs created this way as unaligned context. One example of a situation where a global alignment is preferred over a local alignment is when there are multiple, but disjoint, segments of aligned sequence. Examples when a global alignment is a good choice: 1) aligning a CDS or mRNA sequence to a gene that contains introns; 2) aligning two sequences that differ because of the presence of large insertions, such as might be caused by transposable elements. In both cases, a local alignment is less likely to reflect the full alignment, especially if the lengths of the unalignable inclusions are long relative to the gap extension penalty.
Note: When two sequences are nearly identical (check by performing a multiple alignment and consulting the Distance view), all pairwise methods should work equally well.
- Use the drop-down menu(s) to choose the Substitution matrix and related options, if any
- Specify the Gap open penalty, the amount deducted from the alignment score for each gap in the alignment. Gaps of different sizes carry the same penalty. Default is 10.
- Specify the Gap extension penalty, the value which will be deducted from the alignment score after first multiplying it by the length of gaps. Longer gaps have a greater penalty than shorter gaps. Default is 1.
- If you wish to specify the length of the smallest perfect match of contiguous bases/residues to consider in building an alignment, check the Require minimum word match box and enter a value. The default is unchecked; if checked, the default value is 7.
- Press OK.
During the alignment, MegAlign Pro displays a progress window. In most cases, this will appear and disappear too suddenly to notice it. In the cases of longer alignments, you can interrupt the alignment, if necessary, by clicking its Cancel button or view a console window showing the start time and progress of the alignment by clicking its Show Console button.
If an alignment finishes successfully, a Pairwise view opens. If an alignment fails, you will receive a message with recommendations on how to obtain a successful alignment (e.g., by modifying options or choosing a different alignment method).
- (optional) If the Console is not already open, and you wish to view alignment statistics and other information there, select View > Console.
Need more help with this?
Contact DNASTAR


