When you predict antibody interactions with NovaFold Antibody, the second wizard screen is Options. During the prediction step, options specified in this screen are applied to all jobs listed in the Sequences screen.
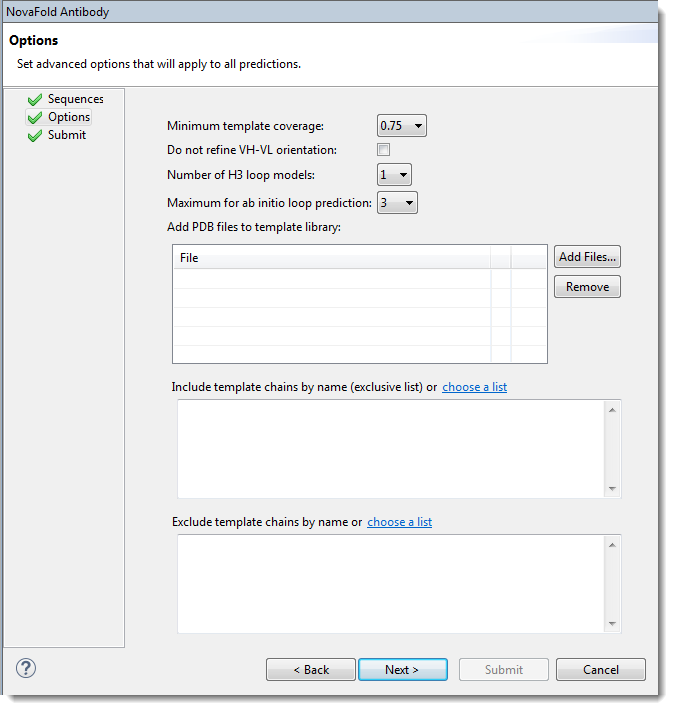
Edit or enter information in this wizard screen as desired. Or you can accept the defaults as they are, and simply press Next to continue to the Submit screen.
- Minimum template coverage – To ensure the selected template exceeds a particular fractional coverage, you may specify a non-zero value using the drop-down menu. This compels NovaFold Antibody to locate a template with both variable and constant domains.
- Do not refine VH-VL orientation – By default, NovaFold Antibody optimizes the rigid-body orientation between the light and heavy antibody chains to remove atomic clashes if they were introduced during the modeling process. To skip the optimization step, check this box.
- Number of H3 loop models – The H3 loop is the generally the hardest region of the antibody structure to predict. NovaFold Antibody offers a template-based approach that uses a machine learning model to choose the best templates for the H3 loop. To specify how many results models to output, choose a value from the drop-down menu.
- Maximum for ab initio loop prediction – To specify a cutoff for switching from ab initio "distance guided" prediction of the H3 loop to a template-based prediction. Example: The default setting of ‘3’ means loops of length 3 or shorter would use the ab initio methodology, while loops longer than 3 residues would be built with the template-based approach.
- Add PDB files to template library – Press Browse to nominate a local 3D structure to use as a template in the modeling prediction, along with other templates selected by NovaFold Antibody. Select a text file representing a single protein chain in standard PDB format. If you need to remove a file from the list, select it and press Remove.
- Include template chains by name (exclusive list) – To define one or more chains from a PDB structure as the template(s) to use in the modeling prediction, type the information into the text area. Each line in the file should represent one template using the format [PDB ID][CHAIN ID][HEAVY/LIGHT]. The PDB ID is four characters, and starts with a number, followed by three letters or numbers. The CHAIN ID is case sensitive and is typically a single character. Examples:
1IGT:A Heavy
1IGT:A Light
Alternatively, press choose a list to open a browser where you can navigate to a local file containing this information.
- Exclude template chains by name – To exclude certain templates from the library (i.e., to prevent these templates from being considered) during structure prediction. Templates can be excluded by name, using the following format: [PDB ID][CHAIN ID]. Example: 1wor:A. A percent sequence identity can be specified at the end, if desired. For example, 1wor:A 70 would specify a 70% sequence identity cutoff. If no number is specified, the percent sequence identity is assumed to be 100%. An asterisk (*) may be used to designate any chain in the PDB file. For example, 1wor:*.
Alternatively, press choose a list to open a browser where you can navigate to a local file containing this information.
Click Next to proceed to the Submit screen, or Back to return to the Sequences screen.
Need more help with this?
Contact DNASTAR


