Featured Post
Lasergene 17.6 Release Notes
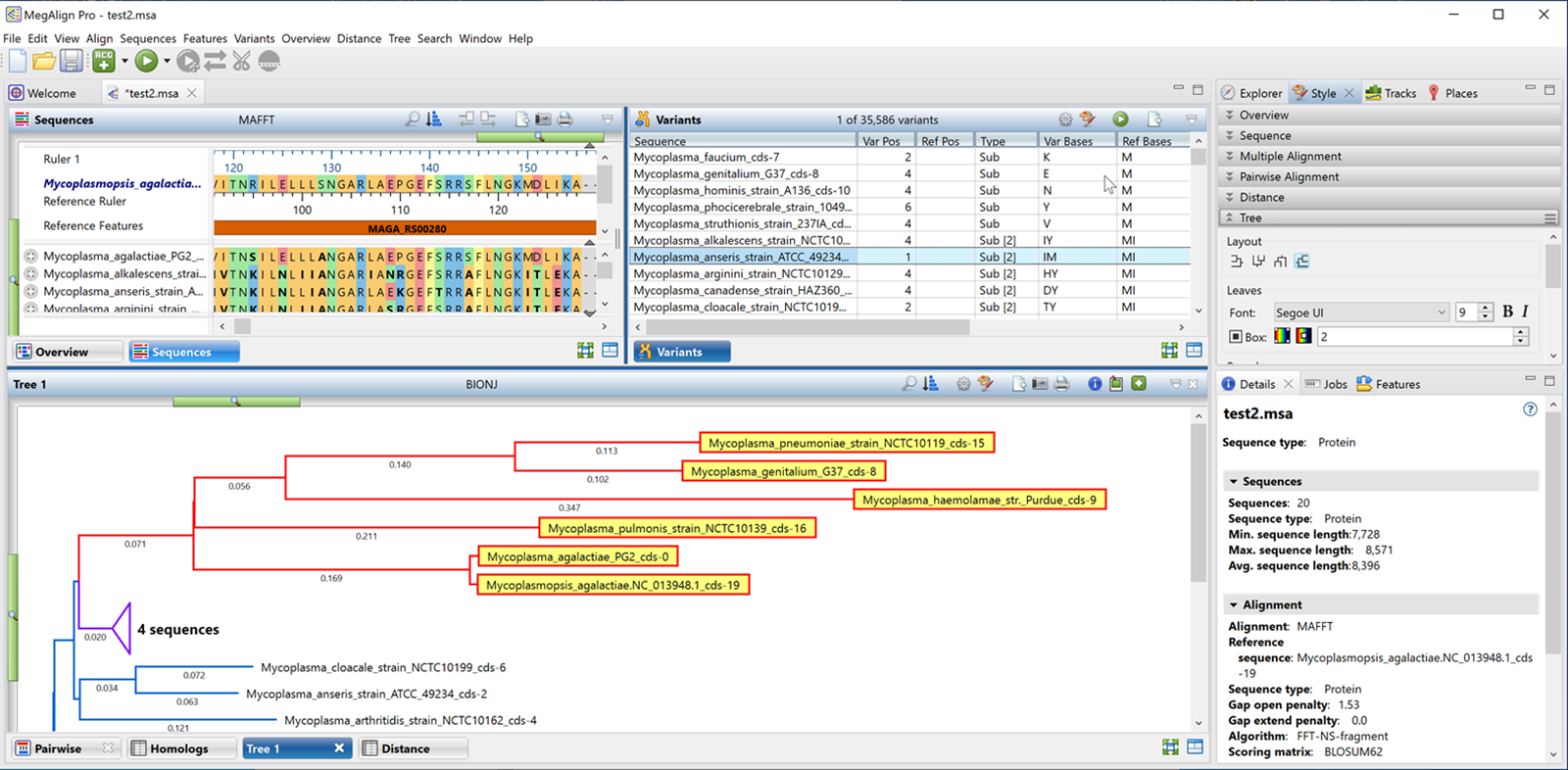
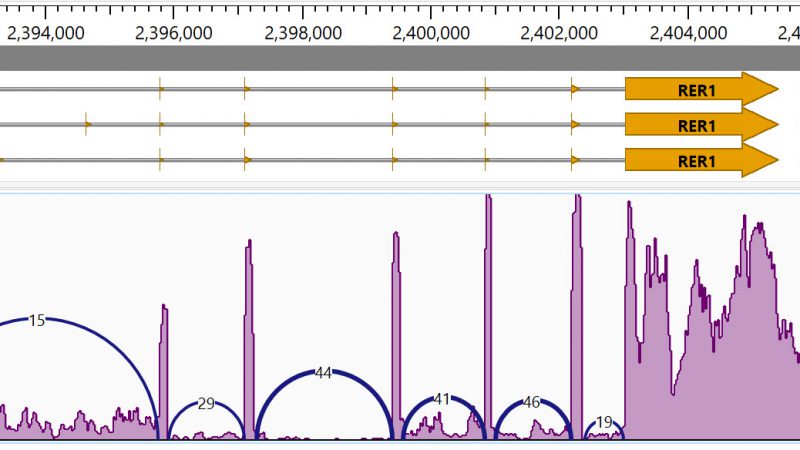
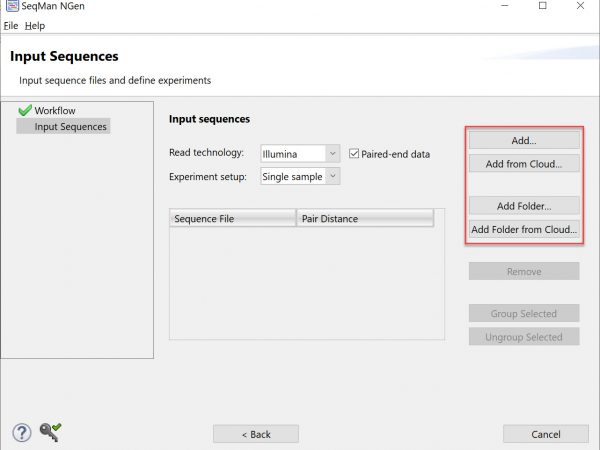
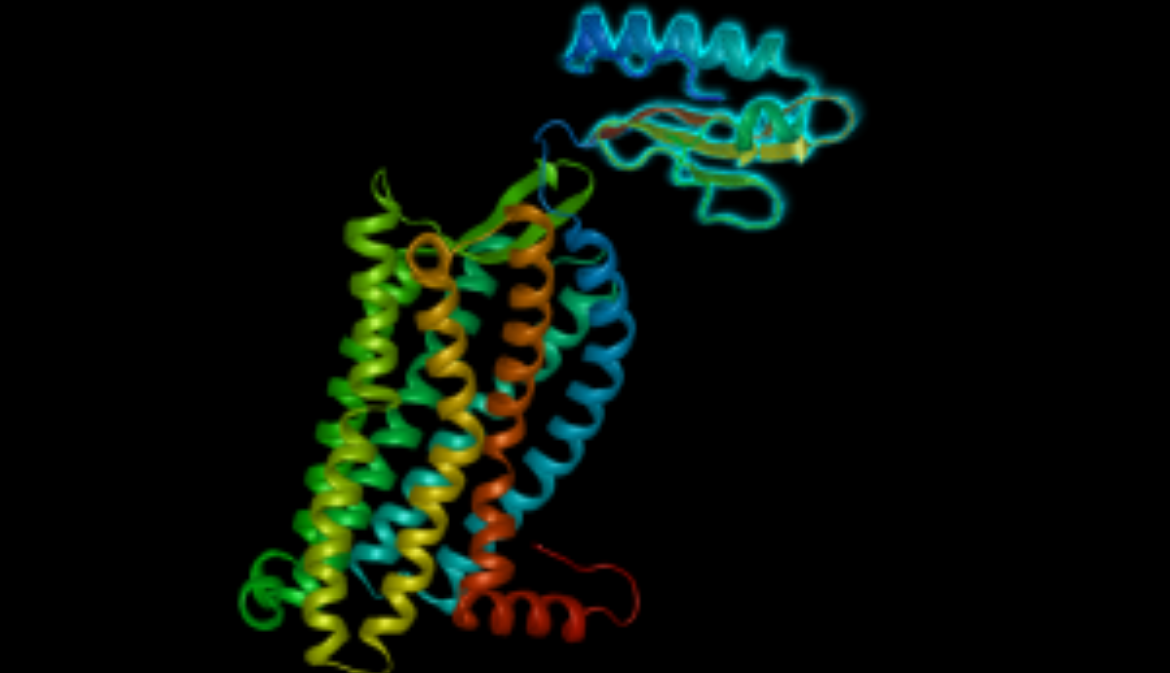
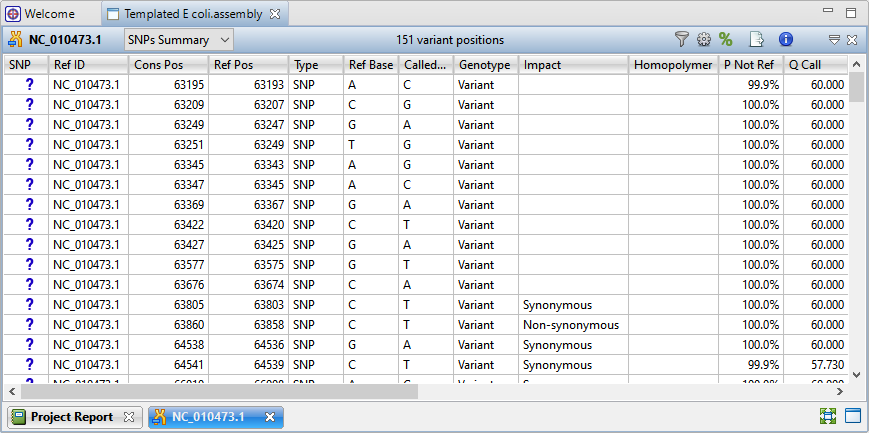
Lasergene has been updated to version 17.6 with remarkable usability improvements for protein analysis in Protean 3D, and the addition of a new Nova workflow that integrates the AlphaFold-multimer method. Additionally, MegAlign Pro has...
featured postRecent Posts
An Integrated Solution for De Novo Transcriptome Analysis