If your vector sequences lack some annotations, or contain incomplete or obsolete versions of annotations, the following procedure lets you effortlessly copy features from a curated Feature Library, or from your own custom features, to the query sequences. Conversely, if you prefer the query annotations to the library versions, you can automatically update the Feature Library version to match the annotations in the query sequences.
Before following the steps below, you may wish to create a custom feature database, specify the features to use in auto-annotation, or specify the appearance of features in the Feature Library Manager and Annotation Results window. These preliminaries are entirely optional.
To auto-annotate a vector sequence from the installed Feature Library and/or your own custom feature database:
- (optional) If you have created multiple “User” databases (e.g. for different organisms), you must change one of the databases to the default library name before proceeding. To do this, navigate to C:\Users\UserName\AppData\Local\DNASTAR/UserFeatures.library (Win) or ~/Library/Preferences (Mac) and temporarily rename the file UserFeatures.library.
- Use either of the following methods to specify which vector sequence(s) to auto-annotate:
- Open the desired sequence in the Document window.
- Open a cloning project in the Project window. Expand the Vectors folder and select one or more files or folders.
- Open the desired sequence in the Document window.
- Choose Features > Annotate Sequence. In the ensuing Annotation Parameters dialog, specify the thresholds for calling a match, or check Perfect matches only to effectively set each value to 100%. By default, SeqBuilder Pro calls a match when a feature sequence matches at least 95% of bases in the query sequence (Minimum match percentage) and overlaps the query sequence by 98% (Minimum coverage percentage). After making the desired selections, press OK.

The Annotation Results window opens. If the query sequence was already up-to-date, a message will appear: “No new features were found.” In this case, click OK to exit from the annotation procedure. Otherwise, the Annotation Results window will populate.

The sortable table at the bottom of the window will likely contain both checked and unchecked rows. A row with a checkmark in its leftmost column represents an annotation that is absent in the query sequence. A row without a checkmark represents an annotation that already exists in the query sequence. The version from SeqBuilder Pro’s plasmid Feature Library may be more accurate or up-to-date than the annotation currently in the query sequence, so you may also wish to examine these entries. See The Annotation Results Window for detailed information about the contents of each pane of the window.
- Click on a feature row to make it the active feature and to populate the rest of the window with information relevant to that feature.
- Use evidence in this window (e.g., match quality, shared endpoints, etc.) to determine whether or not to copy a feature to the query sequence. For instance, you may wish to copy the feature if the plasmid database version is more complete, or if it represents a new feature that does not yet exist in the query sequence.
Example:
An unchecked row was selected in the annotation match table at the bottom of the window. The Matched Feature / Putative Duplicate Feature panes (below) reveal that both features cover an identical range, good evidence that they are duplicates. The feature in the query sequence is named Km( R ), while the plasmid Feature Library calls it KanR but refers to “Kanamycin kinase” and “Geneticin®” in the /note and /function fields, hinting further at a possible match.
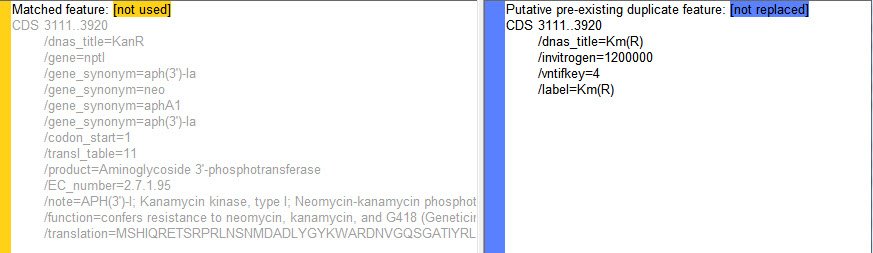
As it happens, Invitrogen/Life Technologies’ Geneticin® is the antibiotic also called G418. It is an aminoglycoside related to Gentamicin. The enzyme encoded by this gene confers resistance to Geneticin as well as the related antibiotics Neomycin and Kanamycin. So Km( R ) and KanR are indeed the same gene. However, the plasmid Feature Library version on the left contains considerably more detailed information, and you will probably want to use this information to overwrite the feature currently in the query sequence.
- To copy checked features from the Feature Library to your query sequence(s) (or vice versa, for rows in which Replace has been specified), click on the Accept Checked tool (
).If you instead wish to exit without copying any features, click the Cancel tool (
).
- If you copied any features to the query, we recommend immediately using File > Save Project to save them. Otherwise, if you later close the project without saving, any changes will be lost.
Need more help with this?
Contact DNASTAR



 ).If you instead wish to exit without copying any features, click the Cancel tool (
).If you instead wish to exit without copying any features, click the Cancel tool (  ).
).