To show/hide sets of enzymes using the Enzymes tool:
To apply groups of enzymes to the sequence based on the frequency of cuts, type or class, site complexity, or overhang compatibility, click the Enzymes tool ( 
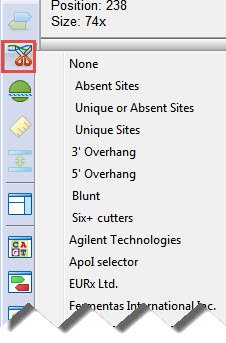
If desired, additional sets of enzymes can be applied to intersect with those in the first selector. When selecting subsequent sets of enzymes, hold down the Ctrl, Alt, or Shift keys (Windows) or Cmd, Ctrl, Option, or Shift (Macintosh.
Once restriction enzyme(s) have been applied to the Sequence view, click and hold on a restriction enzyme label in this view to show the cut sites, represented by thin blue lines. Or do the same while pressing the Alt (Win) or Option (Mac) key to show the cut represented by bold black arrows, and to display the recognition sequence.

To hide an individual enzyme:
Right-click on the enzyme and choose whichever command is available: Hide Enzyme or Hide Selected Enzyme. Alternatively, use the procedure below.
To show/hide individual enzymes using the Enzymes panel:
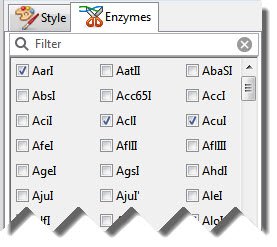
- Open the Enzymes panel by clicking on its tab or by clicking the Enzymes tool (
) and choosing Pick Specific Enzymes.
- In the Enzymes panel, search for enzymes of interest using the responsive search bar at the top.
- To filter the selection to enzymes beginning with particular letters, start typing the enzyme name.
- To remove a filter, click the ‘x’ at the right side of the search bar.
- To filter the selection to enzymes beginning with particular letters, start typing the enzyme name.
- Check the boxes next to enzymes you wish to display and remove check marks from enzymes you wish to hide.
The Details panel will list any restriction enzymes that are suppressed due to very high cut frequency.
- (optional) To predict enzyme cuts for a sequence with ambiguous bases, check the Show Potential Cuts at Ambiguous Bases box under the enzyme list. This box is unchecked by default.
Potential cuts occur when the enzyme recognition sequence and the template sequence are both ambiguous, as can happen when examining a back-translated protein. If the ambiguous residues have a non-empty intersection, then a cut is possible. However, if the intersection does not cover the whole ambiguous set, the cut is not certain. With a fully characterized sequence, all cuts are certain, so there will be no potential cuts.
When Show Potential Cuts at Ambiguous Bases is checked, any restriction enzymes so detected will appear in gray, rather than in blue (Minimap) or black (other views). If you edit the sequence to remove some of the unmatched ambiguity, some of these gray sites will turn black to reflect that the recognition sequence pattern now matches the sequence pattern. Click here to see an example of how this option is used.
Need more help with this?
Contact DNASTAR



