In this tutorial, you will look for a single enzyme capable of excising a histone coding feature from a sequence named X05543.1.
- To open the sequence, follow the tutorial Try it! – Open an Entrez sequence by accession number.
You’ll need an enzyme that cuts in just two places. So you’ll start by creating and applying a frequency selector that displays enzymes that cuts the sequence in a maximum of two places:
- Select Enzymes > New Selector. The Enzyme Selector Manager appears. New Selector is highlighted in the Selector list and the Name is Untitled 1.
- In the Name box, type Two cuts max.
- From the Type drop-down menu, select Frequency.
- Overwrite the number 1 in the Max box with the number 2. Leave all other parameters at their default settings.
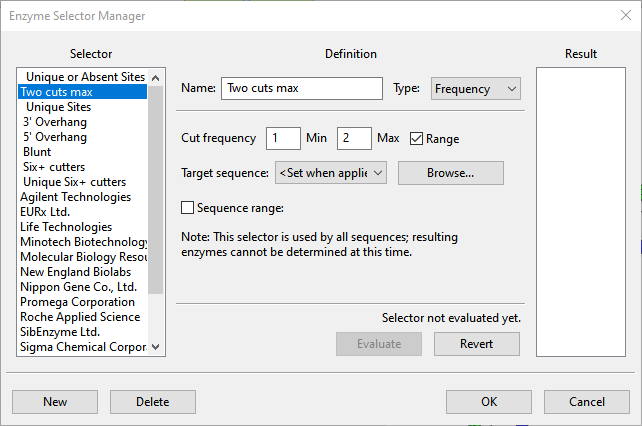
- Click OK to create the new selector and return to the main view.
- Click on the Enzymes tool (
) on the upper left of the window, then choose the new selector Two cuts max to display only the restriction enzymes that will cut this sequence one or two times.
Many restriction sites remain, so you can now use the Minimap view to find the best restriction enzyme for the job:
- In the Circular view (top) or Features view (bottom), single-click on the feature Histone H2B-1 to select the feature in all views. Histone H2B-1 is a CDS feature that was included in the original Genbank-formatted sequence.
- At the bottom of the window, click the Minimap tab to move to that view. The Minimap is scaled to show the whole sequence in a single line. In this view, restrictions enzyme names appear as individual rows in the left-hand column. Their corresponding cut sites are shown in the same row on the right.
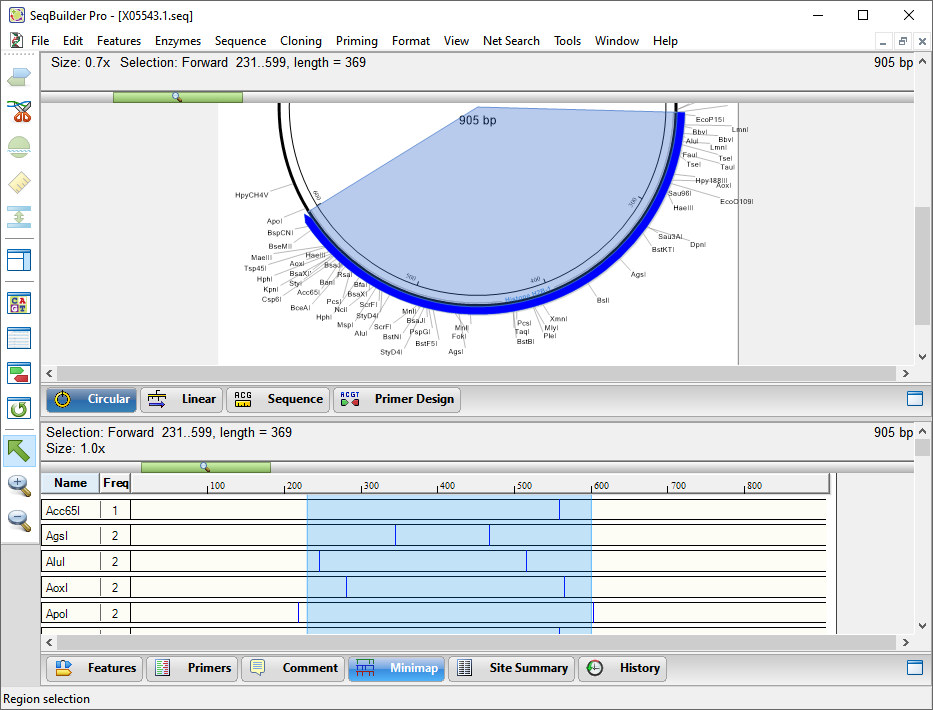
- At the top of the Minimap view, click on the ruler to sort the enzymes by which cut closest to the selection

The restriction sites are sorted so that those cutting closest to and outside the feature appear near the top, those cutting further away from the outside appear further down, and those cutting inside the feature’s range appear at the bottom. Note that the ApoI restriction site cuts just to the left and right of the histone gene, and thus may be used for excising the gene.

- To remove all of the currently-applied enzymes from the sequence, click the Enzymes tool (
) and choose None.
- To apply only the ApoI enzyme, click the tool again and choose Pick Specific Enzymes. This opens the Enzymes panel on the right. Check the box next to ApoI.
This is the end of this tutorial topic.
Need more help with this?
Contact DNASTAR



 ) on the upper left of the window, then choose the new selector Two cuts max to display only the restriction enzymes that will cut this sequence one or two times.
) on the upper left of the window, then choose the new selector Two cuts max to display only the restriction enzymes that will cut this sequence one or two times.